Mini Review
Advances in research of the structural gene characteristics of the aflatoxin biosynthetic gene cluster

Qi-Zhang Li and Zhu-Mei He*
Guangdong Provincial Key Laboratory for Aquatic Economic Animals, School of Life Sciences, Sun Yat-Sen University, Guangzhou 510275, People’s Republic of China
*Address for Correspondence: Prof. Zhu-Mei He, School of Life Sciences, Sun Yat-Sen University, Guangzhou, People’s Republic of China, People’s Republic of China, Tel.: +86 20 84113065; Fax: +86 20 84036551; Email: lsshezm@mail.sysu.edu.cn
Dates: Submitted: 22 October 2018; Approved: 30 October 2018; Published: 31 October 2018
How to cite this article: Li Q, He Z. Advances in research of the structural gene characteristics of the aflatoxin biosynthetic gene cluster. J Plant Sci Phytopathol. 2018; 2: 068-082. DOI: 10.29328/journal.jpsp.1001022
Copyright License: © 2018 Li Q, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Aflatoxins; Structural genes; Aspergillus flavus; Aspergillus parasiticus; promoter
Abstract
Aflatoxins, produced by Aspergillus spp., are strongly toxic and carcinogenic fungal secondary metabolites. Aflatoxin biosynthesis is a complex process and involves at least 30 genes clustered within an approximately 75 kB gene cluster. In this paper, we reviewed current status of the researches on the characterized structural genes involved in aflatoxin biosynthesis and their roles in aflatoxin-producing fungi, especially in A. flavus and A. parasiticus, which will improve our understanding of the mechanism of aflatoxin biosynthesis and regulation and provide reference for further study.
Introduction
As early as 1960, animal poisoning incidents happened in England due to a toxin produced by Aspergillus flavus, and this toxin is called aflatoxins (AFs) later. Aflatoxins produced by Aspergillus spp. are secondary metabolites which have strongly toxicity and carcinogenicity, and an obviously harmful effect on liver [1]. The agricultural contamination caused by aflatoxins is a worldwide problem and a potential threat to animal and human health [2,3]. A. parasiticus and A. flavus are two major aflatoxin producers, and their aflatoxin biosynthesis pathway genes are highly homologus and orders are similar [4]. The aflatoxin production involves a series of complex enzymatic reactions. Every enzyme is encoded by a corresponding structural gene. Therefore, a research on these structural genes directly reflects the mechanism of aflatoxin biosynthesis. This is an important and necessary process to further study the mechanism of synthesis and regulation of aflatoxin and its prevention and control. In this paper, on the basis of previous reviews and research papers [5-9], we mainly focus on the characterizations and roles of structural genes involved in aflatoxin biosynthesis in A. flavus and A. parasiticus and provide reference for more in-depth studies.
Overview of aflatoxin biosynthesis
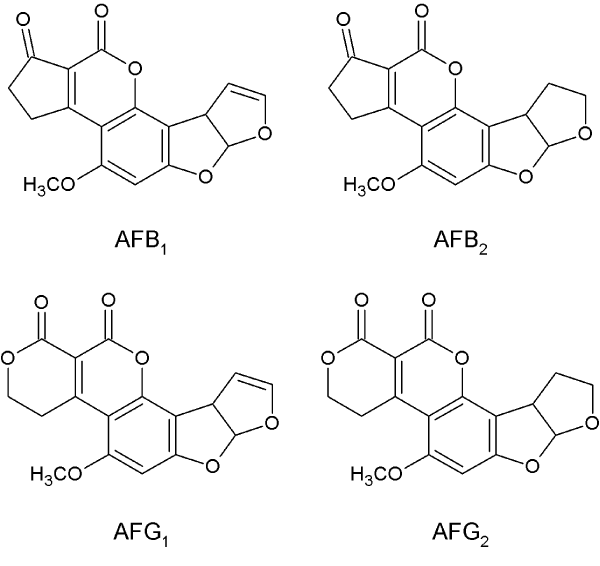
Four different types of aflatoxins are biosynthesized by Aspergilli. Based on different fluorescence under ultraviolet light, these aflatoxins are divided into two groups: one showing blue fluorescence is called B-group aflatoxins, containing AFB1 and AFB2; and the other exhibiting green is called G-group aflatoxins, including AFG1 and AFG2. Moreover, the four aflatoxins are all different in structure (Figure 1). G-group aflatoxin (AFG1 and AFG2) differs from B-group (AFB1 and AFB2) by having an extra oxygen atom in the A-ring; while, 1-group aflatoxin (AFB1 and AFG1) contains a dihydrobisfuran ring and 2-group aflatoxin (AFB2 and AFG2) contains a tetrahydrobisfuran ring [10]. Different Aspergilli produces different kinds of aflatoxins. For example, A. flavus mainly produces B-group aflatoxins, while A. parasiticus produces both B- and G-group aflatoxins. It’s important to note that AFB1 is the most toxic mycotoxin among the four aflatoxins [1]. In addition to aflatoxins, these fungi produce other secondary metabolites as well, such as cyclopiazonic acid, kojic acid, etc. [11,12].
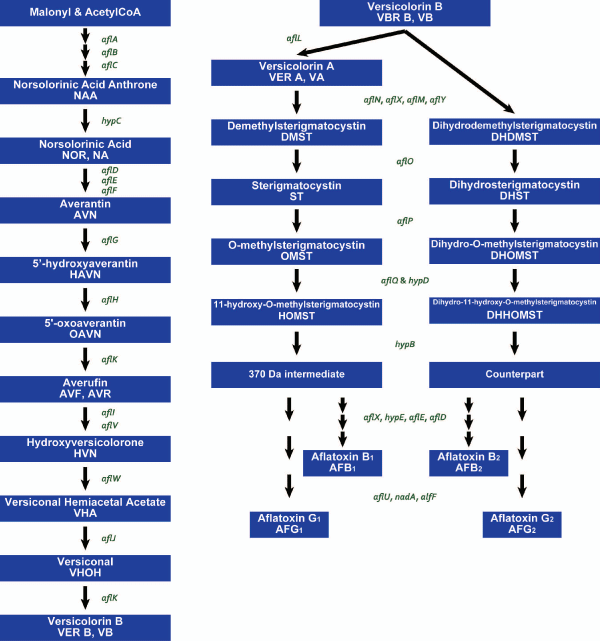
The aflatoxin biosynthesis is complex and involves more than 20 enzyme-catalyzed reactions (Figure 2) [5,8,13]. At least 30 aflatoxin pathway genes are clustered within a 75 kB region of the genome on chromosome III (Figure 3) [6,7]. In the aflatoxin cluster, aflF is at the proximal end of the cluster and nadA is at the distal end. In the 5’ end of the cluster sequence, there is no identifiable open reading frame (ORF) in an approximately 2 kB DNA region and a sugar utilization gene cluster is present in the 3’ end [7]. There are two regulator genes involved in aflatoxin gene cluster and regulating aflatoxin biosynthesis [5].
Structural genes in aflatoxin biosynthesis
Aflatoxin biosynthesis involves a set of enzymatic reactions which need a variety of enzymes encoded by at least 30 genes. These genes encode two fatty acid synthases, a polyketide synthase, eleven oxidoreductases, seven cytochrome P450 monooxygenases, one cyclase, one esterase, two O-methyltransferases and other enzyme.
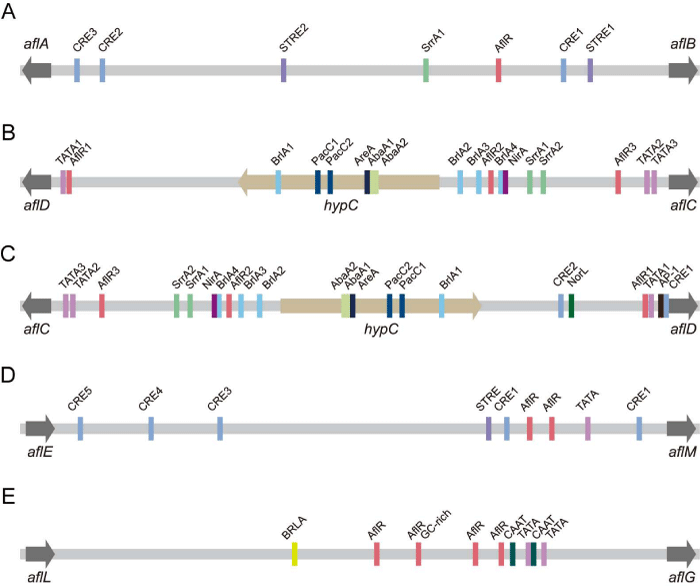
Fatty acid synthase (FAS)-encoding genes: aflA and aflB, two fatty acid synthase encoding genes, are involved in the production of norsolorinic acid anthrone (NAA) from hexanoate produced by acetyl-CoA at the beginning of aflatoxin biosynthesis [14-17]. aflB cloned from A. parasiticus by genetic complementation contains four exons and three introns, and encodes a 7.5 kb transcript of FAS β subunit [6,18]. Besides aflB, the other FAS gene, aflA, which encodes FAS α subunit and contains three exons and two introns, is also found by sequence analysis [6]. The 701 bp of intergenic region between aflA and aflB possesses several elements, such as an atypical AflR-like binding motif or probably two overlapping motifs, three cAMP response element (CRE)-like motifs, two stress-response element (STRE) motifs, a SrrA-binding motif, a CCAAT box and multiple AP-1 motifs [6,19-21] (Figure 4A). On the aspect of protein structure, both AflB and AflA contain some domains. For example, AflB contains acyl transferase (AT) domain, enoyl reductase (ER) domain, dehydrase (DH) domain and malonyl-CoA:acyl-carrier protein (ACP) transacylase (MAT) domain; while, ACP domain, β-ketoacyl synthase (KS) domain, ketoreductase (KR) domain and 4’-phosphopantetheinyltransferase (PPT) domain are found in AflA [8,22].
Polyketide synthase (PKS)-encoding gene: PKS is involved in synthesis of AFB1 polyketide backbone [22,23]. aflC encodes polyketide synthase and is cloned from A. parasiticus [24-26]. This gene has 6.8 kb of genomic DNA, and contains six exons collectively coding for a protein of 2,109 amino acid residues and five introns of 65 bp, 66 bp, 52 bp, 59 bp and 54 bp. In the 1,687 bp of intergenic region between aflC and aflD, the sequence contains many cis-acting elements [19-21,26-28] (Figure 4B). AflR1 have no effect on aflC activity because of the position probably. Both aflD and aflD are required for expression of aflC, although not showing the binding for lacking a C or G at position 8, but having the strongly and moderately competing oligonucleotides 5’-TCG(C/G)(A/T). aflD and BrlA4 help recruit AflR, and aflD is necessary for aflC activity. PacC1 appears to have a positive effect on aflC expression at acid pH and a negative effect at basic pH. Additionally, two AbaA sites, one AreA site, one NirA site, 15 CRE-like motifs, two SrrA-binding motifs, three potential TATA boxes and two inverted CCAAT boxes exist in the promoter sequence. The polyadenylation site of aflC exhibits heterogeneity [26]. Besides the domains recognized by comparing amino acid sequence, more domains are found in AflC: starter unit: ACP transacylase (SAT) domain, KS, MAT, product template (PT), ACP and thioesterase/Claisen cyclase (TE/CLC) [22,29-31].
Oxidoreductase-encoding genes: Eleven oxidoreductases are involved in aflatoxin biosynthesis and encoded by corresponding genes. Some genes only participate in one reaction, such as hypC, aflH, etc. While, some involve in more reactions, such as aflD, aflE, etc.
hypC Norsolorinic acid (NA or NOR) is the first stable precursor in aflatoxin biosynthesis [32]. HypC, an anthrone oxidase, catalyzes the introduction of oxygen into activated aryl moieties, which results in the formation of NA (NOR) from NAA [13]. A putative gene hypC encoding HypC is discovered in the region between aflC and aflD in A. parasiticus [6]. It has the length of 542 bp, contains an intron, and ecodes a protein of 139 amino acid residues. Because hypC is located between aflC and aflD, some of the elements mentioned above which located between aflC and hypC may influence the transcription of hypC (Figure 4B). hypC is predicted to encode approximately 17 kDa proteins and has a catalytic active site Q-L-X-X-Q-W-S-R-I-F-Y [13].
aflD NADPH-dependent NA (NOR) ketoreductase (alcohol dehydrogenase) encoded by aflD catalyzes the conversion of 1’ keto group of NA (NOR) to 1’ hydroxyl group of averantin (AVN) and may be involved in the formation of AFB1 [13,33,34]. aflD transcript is 1.25 kb in length, and contains four exons collectively coding for a 29 kD protein of 294 amino acid residues and three introns of 52 bp, 58 bp and 64 bp [35]. Three transcriptional start sites, two of which are major and one is minor, are located at upstream from the ATG. The polyadenylation site locates approximately 215 bp downstream of translation termination codon. The promoter contains the same elements mentioned in the intergenic region between aflC and aflD (Figure 4C). Multiple transcription factors are required for maximum activity of aflD promoter. AflR1, which has no effect on aflC activity, dose not bind with AflR in EMSA because the A/T at position 5 is replaced by a C, but is necessary, not sufficient for maximum transcriptional activation; while aflD is required for expression of both aflC and aflD [20,27,28,36]. NorL appears only in aflD promoter and is necessary for maximum transcriptional activation in vivo [36]. Two CREs are identified between aflD and hypC, and only CRE1 is functional although they have only one nucleotide difference [20,36]. An AP-1-like site is located at 12 nucleotides upstream from CRE1, and both of them form composite regulatory element [21]. A TATA box is required for maximum transcriptional activation in vivo [20,36]. PacC and BrlA sites do not affect aflD transcriptional regulation, and may not be required for transcriptional activation [36]. A negative regulator presents in the region from position -298 to position -332 by deletion analysis. AflD contains some conserved sequences, such as Y-G-V-S-K-L-A-A-N-Y-M, G-X-G-X-X-L and Y-L-V-T [35]. Additionally, it has high identity and similarity to these of Flavobacterium sp. N-acyl-D-mannosamine dehydrogenase and Streptomyces violaceoruber PKS.
Figure 4: Schematic diagram of cis-acting elements of gene promoter. A, cis-acting elements in the intergenic region between aflA and alfB. B and C, cis-acting elements in the intergenic region between aflC and alfD. D, cis-acting elements in the intergenic region between aflE and alfM. E, cis-acting elements in the intergenic region between aflL and alfG.
aflE and aflF aflE and its homologous gene aflF encode aryl alcohol dehydrogenases [37]. AlfE participates in the formation of AVN and oxidization of AFOH which is yielded in norA disruptant cultures back to AFB1 [13]. aflE is cloned from A. parasiticus using a monoclonal antibody raised against NA (NOR) reductase [38]. An additional copy of aflE, aflE2, is identified by Southern blot analysis, and this may explain higher-level stable aflatoxin produced in A. parasiticus than that in A. flavus which has only one copy. A single ORF of 1,167 bp encodes a 43.7 kDa protein of 388 amino acids. Its promoter region has an AflR-binding motif, a TATA box and a CAAT box [27,38]. The polyadenylation site of aflE transcript is located at nucleotide 1500, and a polyadenylation signal sequence is located 25 bp upstream. aflE and its gene product from A. parasiticus share high degree of identity with its homolog from A. flavus at both the nucleotide and amino acid levels [6,38]. However, they show a low identity to aflD and AflD, respectively [8]. Moreover, there are high homology between aflE and an aryl-alcohol dehydrogenase (aad) gene from Phanerochaete chrysosporium or their products [38]. The amino acid sequence of AflE has an adenine nucleotide binding motif G-X-G-X-X-G and one N-glycosylation site N-X-T.
Besides participation of AVN formation, AflF is responsible for the final oxidation step in AFGs formation [33]. AflF catalyzes an intermediate given from the rearrangement and decarboxylation of the NadA reduced 386 Da intermediate to produce an ester which could be de-esterified by esterases, then this product upon dehydration would yield AFGs. aflF at 5’ end of the gene cluster is cloned by chromosome walking from A. parasiticus [39]. The ORF of aflF consists of 1,146 nucleotides encoding a protein of 382 amino acid residues and no intron is found. However, a portion of aflF is deleted in A. flavus [40]. In the promoter, there is an AflR-binding motif [6,39]. aflF dose not show significant homology with aflE at nucleotide level, but their products share 68% similarity at amino acid level [7].
aflH AflH, a typical short-chain alcohol dehydrogenase, is involved in dehydrogenation of 5’-hydroxyaverantin (HAVN) to 5’-oxoaverantin (OAVN) [41,42]. AflH is a homodimer composed of identical 28 kDa subunits and the native molecular mass is estimated to be 60 kDa [43]. aflH is cloned by chromosome walking and its transcript has approximately 1 kb with no intron [41]. There are two copies of aflH gene, aflH and aflH1, in A. parasiticus SU-1, but only one in A. parasiticus SRRC 2043. Eight nucleotide substitutions presenting in aflH1 results in five amino acid mutations, in which the replacement of G by C in the glycine-rich loop may affect the binding of NAD+-NADP+. aflH promoter region contains an AflR-binding motif [27]. AflH contains two conserved functional motifs, a glycine-rich loop motif G-X-X-X-G-X-G and a motif Y-X-X-X-K [41]. aflH genes or their products in A. flavus and A. parasiticus share no significant homology at either nucleotide level or amino acid level [5].
alfI AflI may catalyze the oxidation of the hydrated intermediate catalyzed by aflV-encoded product, in which aflI-encoded product catalyzes alcohol oxidation of hydroxyversicolorone (HVN) precursor and the ring closure is catalyzed to form the hydroxyfuran [13]. It is reasonable to hypothesize that AflI may form a complex with AflV. aflI is cloned by complementation, and there is only one copy in the A. parasiticus genome [4,44]. aflI consists of 858 bp with no intron and encodes a 30.9 kD protein of 285 amino acid residues. An intergenic region between transcripts of aflO and aflI has only 34 bp, and no TATA motif or AflR-binding site is found [6,44,45]. However, the region at positions -55 and -150 to the translation start contains two TATA boxes. The sequences of aflI and its product from A. parasiticus share very high identity to these from A. flavus and A. sojae either at nucleotide level or at amino acid level. Sequence comparison suggests that the key amino acid at 179 may be responsible for the enzymatic activity.
alfW In the convention of HVN to versiconal hemiacetal acetate (VHA), aflW-encoded HVN monooxygenase catalyzes Baeyer-Villiger reactions [46]. An oxygen atom is inserted into a C-C bond adjacent to the carbonyl group of an aliphatic or alicyclic ketone on the side chain. aflW contains a coding sequence of 1,446 bp encoding a 55 kDa protein of 481 amino acid residues and no intron [47]. aflW is adjacent to aflV and they share the promoter, so aflW has the same promoter elements as these of aflV. Although high homology is showed between AflW and monooxygenases in the GenBank database, no characteristic motif is found.
aflX aflX-encoded oxidoreductase catalyzes the reaction of epoxide ring-opening step during the process of versicolorin A (VER A or VA) to demethylsterigmatocystin (DMST) [48], and may also catalyze an oxidative decarboxylation/dehydration to yield a 326 Da metabolite followed by demethylation to giving AFB1 [13]. aflX, located between aflW and aflY, is cloned from A. parasiticus with no intron and encodes a protein of 266 amino acid residues [6,39]. There is a canonical AflR-binding motif in its promoter [39]. The amino acid sequence of AflX shows 27% identity with that of AflI catalyzed one of the steps in the conversion of averufin (AVR or AVF) to HVN [48], and AflX, like AflI, may be a helper protein for AflV [5,13]. AflX possesses a NADH-flavin reductase motif and several conserved sequences, such as A-X-X-G-A-T-G-X-T-G, R-X-X-X-K-L, L-X-S-A and Y-X-D [39,48].
aflM Deoxygenation of aflM-encoded NADH-dependent deoxygenase/ketoreductase is responsible for formation of DMST and dihydrodemethylsterigmatocystin (DHDMST), in which AflM catalyzes the dienone intermediate given by AflX [49]. aflM, the first gene found involved in aflatoxin biosynthesis, is cloned from A. parasiticus by genetic complementation [50,51]. Two copies are found but only one is functional. aflM is located between aflE and hypE with an intergenic region of 811 bp from aflE [38]. The genomic DNA sequence contains three exons collectively coding for a protein of 262 amino acid residues and two introns of 50 bp and 61 bp [51]. The transcription initiation site is at -82 bp from the translation start codon. In the intergenic region between aflE and aflM, there are two AflR-binding motifs, five CRE-like sites, a SrrA-binding motif, a STRE motif, a TATA box and several AP-1 motifs, but no CAAT box motif is found [19-21,27,51] (Figure 4D). No binding of recombinant AflR with the corresponding motif at -182 bp is observed because the A/T at position 5 in the motif is replaced by a C [27]. The polyadenylation motif is located 98 bp downstream from the translation termination codon and polyadenylation site is located 131 bp [51]. The amino acid sequence of AflM shows significant identity with those of some reductases and dehydrogenases [51-54], and contains an adenine nucleotide binding motif [51,55,56].
alfY aflY-encoded Baeyer-Villiger monoxygenase catalyzes the Baeyer-Villiger oxidation of a dienone intermediate formed by epoxidation of the anthraquinone ring of VER A (VA) [49]. The genomic DNA sequence of aflY contains three exons collectively coding for a protein of 495 amino acid residues and two introns [39]. Its promoter region contains a canonical AflR-binding motif. AflY possesses a highly conserved nicotinate phosphoribosyltransferase (NAPRTase) family motif and several highly conserved regions, such as G-F-H-(N/D)-H-X-X-H-(H/Q), G-X-(L/V)-H-P-(L/I/V)-I-(H/N/Q)-(L/I)-X-X-X-X-E, D-F-X-X-X-H, D-D-G-H-X-X-K-X-X-R-A and W-V-R-W-C-G-(E/D)-X-A-W [49].
HypB HypB can introduce an oxygen into the keto-tautomer of 11-hydroxy-O-methylsterigmatocystin (HOMST), followed by rearrangement to the 370 Da intermediate. While some researchers suggest that this oxidantion is catalyzed by aflQ encoded protein [13,33,57]. hypB is located between aflL and aflI and possesses an intron and encodes a 17 kDa protein of 163 amino acid residues [6,13]. In the promoter, there are two AflR-binding motifs [58].
Cytochrome P450 monooxygenase-encoding genes: Although these gene-encoded enzymes participate in redox reaction, they all belong to cytochrome P450 monooxygenase.
aflG AflG catalyzes the hydroxylation of AVN to form HAVN [37]. Yu et al. clones aflG, located between hypD and aflL, from A. parasiticus by sequencing [4,59]. aflG contains three exons collectively assembling a 1,485 bp of coding region encoding a 56.3 kDa protein of 495 amino acid residues and two introns of 46 bp and 67 bp. Transcription start point of aflG is presented 32 bp upstream of the ATG by primer extension analysis [60]. The 367 bp of intergenic region between aflL and aflG contains four AflR-like binding motifs, an BrlA binding site, an GC-rich dyad symmetric inverted repeat sequence, two functional TATA-like elements and two CAAT boxes [27,59,60] (Figure 4E). Besides, negative regulatory elements may be in the region from -196 to -118 since expression is enhanced by trunction [60]. While, the region from -367 to -196 is not required for aflG gene activity and SrrA-binding site is not found [19,21,60]. The amino acid sequence of AflG contains some conserved motifs, such as F-X-X-G-X-X-X-C-X-G, E-X-X-R and A-G-X-X-T [59].
aflV Disruption of StcB encoding a P450 monooxygenase in the sterigmatocystin (ST) gene cluster of A. nidulans leads to the lack of ST and the accumulation of AVR (AVF), suggesting that this enzyme is involved in the conversion of AVR (AVF) [61]. aflV is homologous to StcB, which is required for the similar reaction in aflatoxin biosynthesis. aflV contains three exons and two introns of 67 and 55 bp, and a coding sequence of 1,473 bp encodes a 56.3 kDa protein of 508 amino acid residues [6,47]. A 620 bp of intergenic region between aflV and aflW contains a canonical AflR-binding motif and four possible TATA boxes. AflV contains three highly conserved motifs, F-X-X-G-X-X-X-C-X-G, E-X-X-R and A-X-X-X-T, which are believed to be the active sites.
aflL Versicolorin B (VER B or VB) is desaturated to VER A (VA) by aflL encoded cytochrome P450 monoxygenase/desaturase, and is also a branch point between biosynthesis of 1-group aflatoxins and that of 2-group aflatoxins [62-64]. Disruption of stcL in A. nidulans abolishes the synthesis of ST and results in the accumulation of VER B (VB) [65]. aflL in A. flavus and A. parasiticus is the homolous gene to stcL [GenBank: AF106958] and is presumed to be involved in this reaction [9]. It is located between aflG and hypB, and is transcribed in a direction same as them. The genomic DNA sequence of aflL contains two exons and an intron [6]. Its promoter region possesses an AflR-binding site but SrrA-binding site is not found [19,21].
aflN stcS is found to be involved in the conversion of VER A (VA) to DMST and its homolog in A. parasiticus is aflN, presuming that aflN is involved in the same conversion in aflatoxin biosynthesis [7,66]. AflN catalyzes epoxidation of the B ring of VER A (VA) [49]. aflN is cloned from A. parasiticus by genomic DNA sequencing [39]. The genomic DNA sequence of aflN contains two exons and an intron, whose ORF encodes a protein of 492 amino acid residues. In the promoter, a canonical AflR-binding motif and three SrrA-binding motifs exist [21,39].
aflQ AflQ catalyzes the epoxidation of the A-ring of O-methylsterigmatocystin (OMST) or dihydro-O-methylsterigmatocystin (DHOMST), then NIH shift to HOMST or dihydro-11-hydroxy-O-methylsterigmatocystin (DHHOMST) [33]. HypD may assist AflQ in oxidation of OMST [13,67-69]. aflQ belongs to a new P-450 gene family named CYP64, and they show more than 90% identity either at nucleotide level or at amino acid level in A. flavus and A. parasiticus [70,71]. Both of them contain seven exons and six introns, although the number of nucleotides in the last two introns are different [71,72]. aflQ codes for a 60.2 kDa protein of 528 amino acid residues. In the promoter region, there are an AflR-like binding site, a TATA box and some elements located in the intergenic region between aflQ and aflP [72]. By site-directed mutagenesis and sequence analysis, H at position 400 is critical for the enzyme activity and A at 143 plays a significant role [71]. The difference in key amino acid residues may influence the ability of aflatoxin production between A. parasiticus and A. flavus. AflQ contains three highly conserved regions, heme-binding motif F-X-X-G-X-X-X-C-X-G, hydrogen bond region E-X-X-R and a highly hydrophobic region [71,72].
aflU AflU can catalyze the 370 Da intermediate to give a 386 Da AFG1 precursor [33]. aflU contains five exons collectively coding for a protein of 498 amino acid residues and four introns of 63 bp, 50 bp, 70 bp and 76 bp [7,39]. Its promoter possesses a canonical AflR-binding motif. Like aflF, a deletion of portion of aflU is present in A. flavus, which may lead to fail to the transcription and loss the ability to produce G-group aflatoxin [40]. AflU possesses some conserved regions of P450 enzymes, such as a heme-binding motif, an E-X-X-R motif and a protein transfer groove A/G-G-X-D/E-T-T/S, and a membrane-binding motif P-X-P [39,40].
NadA NadA-encoded product may catalyze the last conversion from NADA and probably the hypothesized DHNADA (dihydroderivative of NADA) to AFG1 and AFG2 through demethylation in the aflatoxin biosynthesis, in which the NADA produced from AflU catalyzed intermediate has a molecular mass of 360, and is converted to produce AFG1 [73]. Ehrlich et al. [33] shows that NadA is unlikely to catalyze the oxidation of the 360 Da intermediate to AFG1, but NadA catalyzes the 386 Da intermediate given from AflU oxidation to produce a 362 Da intermediate, then converted to 360 Da AFG1 precursor probably corresponding to NADA by NorB. nadA is cloned by gene profiling study using microarray, which is firstly believed to be a sular utilization gene encoding a NADH oxidase [74]. It is the end of the aflatoxin pathway gene cluster at 3’ end. The intron assignment for nadA gene and their protein size are different considerably in different strains. For example, this gene from A. parasiticus has two exons collectively encoding a protein of 444 amino acid residues with 48.5 kD and one intron of 61 bp [33,74]. However, the one from S-strain A. flavus AF70 (GenBank accession number AY510453) contains three exons, encoding a protein of 407 amino acid residues, and two introns; while other from L-strain A. flavus AF13 (GenBank accession number AY510451) contains six exons, encoding a protein of 355 amino acid residues, and five introns [33]. In the promoter, there is an AflR-binding site [58]. The NadA from A. parasiticus possesses a conserved old yellow enzyme (OYE)-related FMN binding domain reductases family domain, and some critical amino acids, such as H at 195, N at 200, and Y at 197. [33,75,76]. In addition to the gene structure and protein size, amino acid composition of NadA is difference, although these diversities are not in the other aflatoxin biosynthesis genes [33,74]. Some amino acids are missing or replaced [33,75]. These sequence differences will result in the inactivation of NadA [33,73,74].
Cyclase-encoding gene: aflK-encoded OAVN cyclase is involved in the formation of AVR (AVF) from OAVN, and is also named versiconal (VHOH or VAL) cyclase or versicolorin B synthase which catalyzes the conversion of VHOH (VAL) to VER B (VB) [43,77,78]. aflK gene product catalyzes intramolecular acetal formation among the given 5’-ketone and two hydroxyl groups (3-OH and 1’-OH) from HAVN [43], and also catalyzes the dehydrative cyclization of racemic VHOH (VAL) to optically active VER B (VB) [62,78,79]. This gene is located between aflQ and aflV, and transcribes in a direction same to aflQ with an intergenic region of 511 bp and opposite to aflV. Its 1,985 bp of genomic DNA sequence contains two exons assembling a continuous ORF of 1,932 bp and one intron of 53 bp, and the coding sequence encodes a protein of 643 amino acid residues with a molecular mass of 70,271 Da and an isoelectric point of 5.06 [80]. The molecular mass and isoelectric point estimated by calculation differ from those observed in experiments. The monomeric molecular mass of native AflK which is a dimer is approximately 78 kDa and the isoelectric point is 4.7 ± 0.1 [78,80]. These differences can be attributed to the post-translational modification. The promoter region contains an AflR-binding site, a pyrimidine-rich motif, a TATA box and a CAAT box [20,27,80]. Besides, this region lacks CRE-like motif and dose not bind AftB as well [19-21]. A polyadenylation site at 161 bp after the termination codon does not correspond to the canonical one [80]. In comparison of amino acid sequences, AflK shows high homology to many flavin-dependent oxidases and dehydrogenases. Its amino acid sequence contains three N-linked glycosylation sites N-X-S/T, a G-X-G-X-X-G motif and a calmodulin binding domain (CaMBD) [80-82].
Esterase-encoding gene: A esterase encoded by aflJ involved in the conversion of VHA to VHOH (VAL) has been purified from A. parasiticus [83,84]. An additional copy of aflJ, estA2, whose gene product has 9 amino acid substitutions, is outside the aflatoxin pathway gene cluster but is not expressed for position effect likely [85]. The 1,000 bp of aflJ genomic DNA contains two exons collectively assembling a coding sequence of 945 bp encoding a 34 kDa protein of 314 amino acid residues and one intron of 55 bp. AflJ is an about 60 kDa of isomeric dimer protein consisting of two 32 kDa of monomer [83,84]. The promoter of aflJ contains consecutive two canonical AflR-binding motifs, a SrrA-binding motif, five TATA boxes and one CAAT box [21,85].
O-methlytransferase-encoding genes: There are two O-methlytransferases involved in aflatoxin biosynthesis [86]. The primary structures of the two O-methlytransferase genes and their products exhibit many difference, so that their biological functions are different [45,86]. O-methyltransferase I encoded by aflO catalyzes methylation of 6-hydroxyl group of DMST or DHDMST in order to produce ST or dihydrosterigmatocystin (DHST), then O-methyltransferase II, the other O-methyltransferase encoded by aflP, further methylates 7-OH of ST or DHST to form OMST or DHOMST [86].
aflO The cDNA sequence of aflO has the length of 1,373 bp with three introns of 63 bp, 50 bp and 62 bp, and encodes a 43 kDa protein of 386 amino acid residues [6,45,87]. The transcription start site is at -140 bp upstream before translation start codon. The promoter region possesses an AflR-binding site and three TATA boxes, but no SrrA-binding site is found [19,21,45]. The polyadenylation signal sequence is at 26 bp downstream of the translation termination codon and the polyadenylation site is at 44 bp. AflO contains an SAM binding consensus sequence and a CaMBD [45,82].
aflP The genomic DNA sequence of aflP contains a 1,484 bp of ORF composed of five exons collectively coding for a 46 kD protein of 418 amino acid residues and four introns of 6 bp, 59 bp, 52 bp and 59 bp [88,89]. In the promoter region, there are two AflR-binding sites, nine CRE-like motifs, a STRE motif, two or three TATA boxes and four CAAT boxes but no SrrA-binding site [19-21,27,88]. The nucleotide sequences of aflP from A. parasiticus and A. flavus share more than 97% identity and same as at amino acid level [88]. The native AflP has a leader peptide of 41 amino acid residues, and the mature one consists of 377 amino acid residues which has a molecular weight of 42 kD [89]. AflP possesses a SAM-binding site V-L-(E/D)-X-G-X-G-X-G, a CaMBD and a sequence Y-L-V-T near the N terminus [35,82,88,89].
Other enzyme-encoding gene: hypE encodes a product which can catalyze the 328 Da intermediate from the 326 Da intermediate with an unknown cytochrome P450 enzyme to lose the methyl residue for the production of AFB1 [13]. HypE has a catalytic ethD domain required for ethyl-t-butyl ether hydrolysis [90].
Prospects
Researches of aflatoxins, the highly toxic and carcinogenic fungal secondary metabolites produced by A. flavus and A. parasiticus, have been extensively carried out more than 50 years since 1960s. Complete nucleotide sequences of aflatoxin biosynthesis clusters contained in several aflatoxin-producing varieties including A. flavus and A. parasiticus are already been known [6]. Genes involved in aflatoxin biosynthesis, including structural and regulatory genes, have been reported [5]. With the identification of functions of the genes in the cluster, the mechanism of aflatoxin biosynthesis seems to have been unveiled.
Actually, in addition to structural genes in the cluster, aflatoxin biosynthesis is regulated by other factors, such as regulatory genes, modification, environmental elements, etc. DNA methylation, an epigenetic modification, plays a vital role in the regulation of gene expression in eukaryotes. DNA methyltransferase inhibitor 5-azacytidine (5-AC) can induce a fluffy phenotype and inhibit aflatoxin biosynthesis in A. flavus [91]. Unexpectedly, our results reveal that the DNA methylation level of A. flavus is negligible [92]. In such situations, we believe that DNA methylation does not act to regulate the expressions of the genes involved in aflatoxin pathways and is not a target of 5-AC. Moreover, we find that DmtA, a DNA methyltransferase homologue, is important in regulation of aflatoxin biosynthesis and for A. flavus to adapt to stressful environments and for survival [93]. This increase the complexity in the regulation of aflatoxin biosynthesis. The canonical regulatory genes within the AF gene cluster, aflR and aflS, have been extensively investigated [94]. Beyond them, some other regulatory factors, such as LaeA, Ham, NosA, FarB, CreA, etc., can also influence aflatoxin formation in A. flavus [91,95,96].
In this review, we pay more attention to structural genes. Some details in the whole process of aflatoxin biosynthesis are not very clear, especially in the last steps which may need additional enzyme(s) but has not been identified yet. In order to do that, many more studies have been made in biochemical and molecular mechanisms of aflatoxin biosynthesis. In the process of structural gene cloning, multiple methods are carried out, such as genetic complementation (aflB, aflD, aflI, etc.), reverse genetics (aflE, aflP, etc.), chromosome walking (aflF, aflH, aflQ, etc.), sequence analysis (hypC, aflV, aflN, etc.), etc., in which genetic complementation is considered to be an effective strategy [97]. The functions of the most of these genes have been confirmed by gene knockout, gene complementation studies, or enzymatic studies [7]. Every structural gene can catalyze a specific reaction or multiple reactions in aflatoxins formation. Regulatory genes, such as aflR, and other biotic and abiotic elements, such as carbon source, nitrogen source and bacteria, also affect aflatoxin biosynthesis [5,98]. With the report of genome sequence of A. flavus [99], more information will be obtained to research whether it exists other structural gene(s) and regulatory gene(s) needed in the aflatoxin biosynthesis in or beyond the aflatoxin biosynthetic gene cluster. Therefore, identification of all the structural and regulatory genes involved in the aflatoxin biosynthesis becomes imperative. Using some new strategies including the Next Generation Sequencing technology will gain more knowledge about genetic regulation, signal transduction and interaction with the environment [91]. With more information being summarized, we will have a better understanding of mechanisms of aflatoxin biosynthesis and regulation and its prevention and control.
Acknowledgement
This research was funded by the National Natural Science Foundation of China (Grant No. 31170044 and 31870031), and the Key Project of Natural Science Foundation of Guangdong Province of China (Grant No. 2016B020204001).
References
- Eaton DL, Gallagher EP. Mechanisms of aflatoxin carcinogenesis. Annu Rev Pharmacol Toxicol. 1994; 34: 135-172. Ref.: https://goo.gl/8JAKV7
- >Bennett JW. Mycotoxins, mycotoxicoses, mycotoxicology and Mycopathologia. Mycopathologia. 1987; 100: 3-5. Ref.: https://goo.gl/CmPAS5
- Massey TE, Stewart RK, Daniels JM, Liu L. Biochemical and molecular aspects of mammalian susceptibility to aflatoxin B1 carcinogenicity. Proc Soc Exp Biol Med. 1995; 208: 213-227. Ref.: https://goo.gl/d9y8sT
- Yu J, Chang PK, Cary JW, Wright M, Bhatnagar D, et al. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl Environ Microbiol. 1995; 61: 2365-2371. Ref.: https://goo.gl/QAQBW8
- Yu J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins (Basel). 2012; 4: 1024-1057. Ref.: https://goo.gl/CZ694W
- Ehrlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol. 2005; 99: 518-527. Ref.: https://goo.gl/LQLFdo
- Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004; 70: 1253-1262. Ref.: https://goo.gl/zbda36
- Yabe K, Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2004; 64: 745-755. Ref.: https://goo.gl/zPC4Sf
- Bhatnagar D, Ehrlich KC, Cleveland TE. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2003; 61: 83-93. https://goo.gl/L5eeWW
- Yabe K, Ando Y, Hamasaki T. Biosynthetic relationship among aflatoxins B1, B2, G1, and G2. Appl Environ Microbiol. 1988; 54: 2101-2106. Ref.: https://goo.gl/PLn7Uw
- Trucksess MW, Mislivec PB, Young K, Bruce VR, Page SW. Cyclopiazonic acid production by cultures of Aspergillus and Penicillium species isolated from dried beans, corn meal, macaroni, and pecans. J Assoc Off Anal Chem. 1987; 70: 123-126. Ref.: https://goo.gl/LbmR4V
- Rosfarizan M, Ariff AB, Hassan MA, Karim MI. Kojic acid production by Aspergillus flavus using gelatinized and hydrolyzed sago starch as carbon sources. Folia Microbiol (Praha). 1998; 43: 459-464. Ref.: https://goo.gl/TfxyP6
- Ehrlich KC. Predicted roles of the uncharacterized clustered genes in aflatoxin biosynthesis. Toxins (Basel). 2009; 1: 37-58. Ref.: https://goo.gl/rsMKXe
- Watanabe CM, Wilson D, Linz JE, Townsend CA. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem Biol. 1996; 3: 463-469. Ref.: https://goo.gl/bjsyQ1
- Watanabe CM, Townsend CA. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B(1). Chem Biol. 2002; 9: 981-988. Ref.: https://goo.gl/q7jwaT
- Hitchman TS, Schmidt EW, Trail F, Rarick MD, Linz JE, et al. Hexanoate synthase, a specialized type I fatty acid synthase in aflatoxin B1 biosynthesis. Bioorg Chem. 2001; 29: 293-307. Ref.: https://goo.gl/YfrbLn
- Townsend CA, Christensen SB, Trautwein K. Hexanoate as a starter unit in polyketide biosynthesis. J Am Chem Soc. 1984; 106: 3868-3869. Ref.: https://goo.gl/Ua2fQB
- Mahanti N, Bhatnagar D, Cary JW, Joubran J, Linz JE. Structure and function of fas-1A, a gene encoding a putative fatty acid synthetase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1996; 62: 191-195. Ref.: https://goo.gl/DeXEb9
- Hong SY, Roze LV, Wee J, Linz JE. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiologyopen. 2013; 2: 144-160. Ref.: https://goo.gl/AkvVWj
- Roze LV, Miller MJ, Rarick M, Mahanti N, Linz JE. A novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J Biol Chem. 2004; 279: 27428-27439. Ref.: https://goo.gl/BUJME6
- Roze LV, Chanda A, Wee J, Awad D, Linz JE. Stress-related transcription factor AtfB integrates secondary metabolism with oxidative stress response in aspergilli. J Biol Chem. 2011; 286: 35137-35148. Ref.: https://goo.gl/fXmhTw
- Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci U S A. 2006; 103: 16728-16733. Ref.: https://goo.gl/R1JscB
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, et al. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009; 461: 1139-1143. Ref.: https://goo.gl/95sTJk
- Chang PK, Cary JW, Yu J, Bhatnagar D, Cleveland TE. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B1 biosynthesis. Mol Gen Genet. 1995; 248: 270-277. Ref.: https://goo.gl/b2iYwm
- Trail F, Mahanti N, Rarick M, Mehigh R, Liang SH, et al. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl Environ Microbiol. 1995; 61: 2665-2673. Ref.: https://goo.gl/nkPc5w
- Feng GH, Leonard TJ. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J Bacteriol. 1995; 177: 6246-6254. Ref.: https://goo.gl/M4Kgf7
- Ehrlich KC, Montalbano BG, Cary JW. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene. 1999; 230: 249-257. Ref.: https://goo.gl/93LpKG
- Ehrlich KC, Montalbano BG, Cary JW, Cotty PJ. Promoter elements in the aflatoxin pathway polyketide synthase gene. Biochim Biophys Acta. 2002; 1576: 171-175. Ref.: https://goo.gl/GW2XiA
- Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002; 323: 585-598. Ref.: https://goo.gl/P7gSqS
- Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg Chem. 2008; 36: 16-22. Ref.: https://goo.gl/e7uUnF
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008; 320: 243-246. Ref.: https://goo.gl/XARYms
- Bennett JW, Christensen SB. New perspectives on aflatoxin biosynthesis. Adv Appl Microbiol. 1983; 29: 53-92. Ref.: https://goo.gl/LdqTXq
- Ehrlich KC, Scharfenstein LL Jr., Montalbano BG, Chang PK. Are the genes nadA and norB involved in formation of aflatoxin G(1)? Int J Mol Sci. 2008; 9: 1717-1729. Ref.: https://goo.gl/V8fJv4
- Zhou R, Linz JE. Enzymatic function of the nor-1 protein in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1999; 65: 5639-5641. Ref.: https://goo.gl/8VYKGB
- Trail F, Chang PK, Cary J, Linz JE. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl Environ Microbiol. 1994; 60: 4078-4085. Ref.: https://goo.gl/bvqnoQ
- Miller MJ, Roze LV, Trail F, Linz JE. Role of cis-acting sites NorL, a TATA box, and AflR1 in nor-1 transcriptional activation in Aspergillus parasiticus. Appl Environ Microbiol. 2005; 71: 1539-1545. Ref.: https://goo.gl/YwkVTu
- Yabe K, Nakamura Y, Nakajima H, Ando Y, Hamasaki T. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl Environ Microbiol. 1991; 57: 1340-1345. Ref.: https://goo.gl/T1SZo4
- Cary JW, Wright M, Bhatnagar D, Lee R, Chu FS. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl Environ Microbiol. 1996; 62: 360-366. Ref.: https://goo.gl/VA1iw1
- Yu J, Bhatnagar D, Cleveland TE. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004; 564: 126-130. Ref.: https://goo.gl/MEzEze
- Ehrlich KC, Chang PK, Yu J, Cotty PJ. Aflatoxin biosynthesis cluster gene cypA is required for G aflatoxin formation. Appl Environ Microbiol. 2004; 70: 6518-6524. Ref.: https://goo.gl/Ukixmd
- Chang PK, Yu J, Ehrlich KC, Boue SM, Montalbano BG, et al. adhA in Aspergillus parasiticus is involved in conversion of 5'-hydroxyaverantin to averufin. Appl Environ Microbiol. 2000; 66: 4715-4719. Ref.: https://goo.gl/q8HbAL
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, et al. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 1999; 27: 260-262. Ref.: https://goo.gl/DvrSZ5
- Sakuno E, Yabe K, Nakajima H. Involvement of two cytosolic enzymes and a novel intermediate, 5'-Oxoaverantin, in the pathway from 5'-Hydroxyaverantin to averufin in aflatoxin biosynthesis. Appl Environ Microbiol. 2003; 69: 6418-6426. Ref.: https://goo.gl/uJYiWt
- Yu J, Woloshuk CP, Bhatnagar D, Cleveland TE. Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene. 2000; 248: 157-167. Ref.: https://goo.gl/1LwU74
- Motomura M, Chihaya N, Shinozawa T, Hamasaki T, Yabe K. Cloning and characterization of the O-methyltransferase I gene (dmtA) from Aspergillus parasiticus associated with the conversions of demethylsterigmatocystin to sterigmatocystin and dihydrodemethylsterigmatocystin to dihydrosterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1999; 65: 4987-4994. Ref.: https://goo.gl/oSedyJ
- Yabe K, Chihaya N, Hamamatsu S, Sakuno E, Hamasaki T, et al. Enzymatic conversion of averufin to hydroxyversicolorone and elucidation of a novel metabolic grid involved in aflatoxin biosynthesis. Appl Environ Microbiol. 2003; 69: 66-73. Ref.: https://goo.gl/Zry8nN
- Yu J, Chang PK, Bhatnagar D, Cleveland TE. Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl Microbiol Biotechnol. 2000; 53: 583-590. Ref.: https://goo.gl/cyqcvZ
- Cary JW, Ehrlich KC, Bland JM, Montalbano BG. The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of versicolorin A to demethylsterigmatocystin. Appl Environ Microbiol. 2006; 72: 1096-1101. Ref.: https://goo.gl/CG9Sxb
- Ehrlich KC, Montalbano B, Boue SM, Bhatnagar D. An aflatoxin biosynthesis cluster gene encodes a novel oxidase required for conversion of versicolorin a to sterigmatocystin. Appl Environ Microbiol. 2005; 71: 8963-8965. Ref.: https://goo.gl/iPggiS
- Liang SH, Skory CD, Linz JE. Characterization of the function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1996; 62: 4568-4575. Ref.: https://goo.gl/uUepJY
- Skory CD, Chang PK, Cary J, Linz JE. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl Environ Microbiol. 1992; 58: 3527-3537. Ref.: https://goo.gl/YVtkf2
- Hallam SE, Malpartida F, Hopwood DA. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene. 1988; 74: 305-320. Ref.: https://goo.gl/EPXthu
- Delledonne M, Porcari R, Fogher C. Nucleotide sequence of the nodG gene of Azospirillum brasilense. Nucleic Acids Res. 1990; 18: 6435. Ref.: https://goo.gl/6G4AsR
- Heilmann HJ, Magert HJ, Gassen HG. Identification and isolation of glucose dehydrogenase genes of Bacillus megaterium M1286 and their expression in Escherichia coli. Eur J Biochem. 1988; 174: 485-490. Ref.: https://goo.gl/rUd2k4
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990; 24: 37-66. Ref.: https://goo.gl/1AwzYP
- Kamps MP, Taylor SS, Sefton BM. Direct evidence that oncogenic tyrosine kinases and cyclic AMP-dependent protein kinase have homologous ATP-binding sites. Nature. 1984; 310: 589-592. Ref.: https://goo.gl/R3KUKy
- Udwary DW, Casillas LK, Townsend CA. Synthesis of 11-hydroxyl O-methylsterigmatocystin and the role of a cytochrome P-450 in the final step of aflatoxin biosynthesis. J Am Chem Soc. 2002; 124: 5294-5303. Ref.: https://goo.gl/2ju9Au
- Price MS, Yu J, Nierman WC, Kim HS, Pritchard B, et al. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol Lett. 2006; 255: 275-279. Ref.: https://goo.gl/kmx4hm
- Yu J, Chang PK, Cary JW, Bhatnagar D, Cleveland TE. avnA, a gene encoding a cytochrome P-450 monooxygenase, is involved in the conversion of averantin to averufin in aflatoxin biosynthesis in Aspergillus parasiticus. Appl Environ Microbiol. 1997; 63: 1349-1356. Ref.: https://goo.gl/4emi7c
- Cary JW, Montalbano BG, Ehrlich KC. Promoter elements involved in the expression of the Aspergillus parasiticus aflatoxin biosynthesis pathway gene avnA. Biochim Biophys Acta. 2000; 1491: 7-12. Ref.: https://goo.gl/oapVdn
- Keller NP, Watanabe CM, Kelkar HS, Adams TH, Townsend CA. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl Environ Microbiol. 2000; 66: 359-362. Ref.: https://goo.gl/iwAA8S
- Yabe K, Hamasaki T. Stereochemistry during aflatoxin biosynthesis: cyclase reaction in the conversion of versiconal to versicolorin B and racemization of versiconal hemiacetal acetate. Appl Environ Microbiol. 1993; 59: 2493-2500. Ref.: https://goo.gl/dr19gi
- Bhatnagar D, Cleveland TE, Kingston DG. Enzymological evidence for separate pathways for aflatoxin B1 and B2 biosynthesis. Biochemistry. 1991; 30: 4343-4350. Ref.: https://goo.gl/KMPVYv
- Yabe K, Ando Y, Hamasaki T. Desaturase activity in the branching step between aflatoxins B1 and G1 and aflatoxins B2 and G2. Agric Biol Chem. 1991; 55: 1907-1911. Ref.: https://goo.gl/t6DqxM
- Kelkar HS, Skloss TW, Haw JF, Keller NP, Adams TH. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J Biol Chem. 1997; 272: 1589-1594. Ref.: https://goo.gl/RTJcZ6
- Keller NP, Segner S, Bhatnagar D, Adams TH. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans. Appl Environ Microbiol. 1995; 61: 3628-3632. Ref.: https://goo.gl/c76Qba
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002; 66: 447-459. Ref.: https://goo.gl/BAePgG
- Reiss J. Development of Aspergillus parasiticus and formation of aflatoxin B1 under the influence of conidiogenesis affecting compounds. Arch Microbiol. 1982; 133: 236-238. Ref.: https://goo.gl/KQR3cu
- Wieser J, Yu JH, Adams TH. Dominant mutations affecting both sporulation and sterigmatocystin biosynthesis in Aspergillus nidulans. Curr Genet. 1997; 32: 218-224. Ref.: https://goo.gl/bQdk9g
- Prieto R, Yousibova GL, Woloshuk CP. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl Environ Microbiol. 1996; 62: 3567-3571. Ref.: https://goo.gl/SbGCgC
- Yu J, Chang PK, Ehrlich KC, Cary JW, Montalbano B, et al. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl Environ Microbiol. 1998; 64: 4834-4841. Ref.: https://goo.gl/CAFAPM
- Prieto R, Woloshuk CP. ord1, an oxidoreductase gene responsible for conversion of O-methylsterigmatocystin to aflatoxin in Aspergillus flavus. Appl Environ Microbiol. 1997; 63: 1661-1666. Ref.: https://goo.gl/7Huf4a
- Cai J, Zeng H, Shima Y, Hatabayashi H, Nakagawa H, et al. Involvement of the nadA gene in formation of G-group aflatoxins in Aspergillus parasiticus. Fungal Genet Biol. 2008; 45: 1081-1093. Ref.: https://goo.gl/y6VNYo
- Yu J, Chang P, Bhatnagar D, Cleveland TE. Cloning of a sugar utilization gene cluster in Aspergillus parasiticus. Biochim Biophys Acta. 2000; 1493: 211-214. Ref.: https://goo.gl/u8AkSx
- Fox KM, Karplus PA. Old yellow enzyme at 2 Å resolution: overall structure, ligand binding, and comparison with related flavoproteins. Structure. 1994; 2: 1089-1105. Ref.: https://goo.gl/oEWSf6
- Williams RE, Bruce NC. 'New uses for an Old Enzyme'--the Old Yellow Enzyme family of flavoenzymes. Microbiology. 2002; 148: 1607-1614. Ref.: https://goo.gl/QRPEoA
- Sakuno E, Wen Y, Hatabayashi H, Arai H, Aoki C, et al. Aspergillus parasiticus cyclase catalyzes two dehydration steps in aflatoxin biosynthesis. Appl Environ Microbiol. 2005; 71: 2999-3006. Ref.: https://goo.gl/doux9q
- McGuire SM, Silva JC, Casillas EG, Townsend CA. Purification and characterization of versicolorin B synthase from Aspergillus parasiticus. Catalysis of the stereodifferentiating cyclization in aflatoxin biosynthesis essential to DNA interaction. Biochemistry. 1996; 35: 11470-11486. Ref.: https://goo.gl/A3KPSS
- Lin BK, Anderson JA. Purification and properties of versiconal cyclase from Aspergillus parasiticus. Arch Biochem Biophys. 1992; 293: 67-70. Ref.: https://goo.gl/XHcZKv
- Silva JC, Minto RE, Barry CE 3rd, Holland KA, Townsend CA. Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. Expansion of the aflatoxin b1 biosynthetic gene cluster. J Biol Chem. 1996; 271: 13600-13608. Ref.: https://goo.gl/rjgw9B
- Silva JC, Townsend CA. Heterologous expression, isolation, and characterization of versicolorin B synthase from Aspergillus parasiticus. A key enzyme in the aflatoxin B1 biosynthetic pathway. J Biol Chem. 1997; 272: 804-813. Ref.: https://goo.gl/Vgk1c4
- Juvvadi PR, Chivukula S. Putative calmodulin-binding domains in aflatoxin biosynthesis-regulatory proteins. Curr Microbiol. 2006; 52: 493-496. Ref.: https://goo.gl/Zn1k4d
- Hsieh DP, Wan CC, Billington JA. A versiconal hemiacetal acetate converting enzyme in aflatoxin biosynthesis. Mycopathologia. 1989; 107: 121-126. Ref.: https://goo.gl/vBLdB5
- Kusumoto K, Hsieh DP. Purification and characterization of the esterases involved in aflatoxin biosynthesis in Aspergillus parasiticus. Can J Microbiol. 1996; 42: 804-810. Ref.: https://goo.gl/bUqseb
- Yu J, Chang PK, Bhatnagar D, Cleveland TE. Cloning and functional expression of an esterase gene in Aspergillus parasitcus. Mycopathologia. 2002; 156: 227-234. Ref.: https://goo.gl/8uP3Dv
- Yabe K, Ando Y, Hashimoto J, Hamasaki T. Two distinct O-methyltransferases in aflatoxin biosynthesis. Appl Environ Microbiol. 1989; 55: 2172-2177. Ref.: https://goo.gl/DVHfJM
- Yabe K, Matsushima K, Koyama T, Hamasaki T. Purification and characterization of O-methyltransferase I involved in conversion of demethylsterigmatocystin to sterigmatocystin and of dihydrodemethylsterigmatocystin to dihydrosterigmatocystin during aflatoxin biosynthesis. Appl Environ Microbiol. 1998; 64: 166-171. Ref.: https://goo.gl/fTbHrK
- Yu J, Chang PK, Payne GA, Cary JW, Bhatnagar D, et al. Comparison of the omtA genes encoding O-methyltransferases involved in aflatoxin biosynthesis from Aspergillus parasiticus and A. flavus. Gene. 1995; 163: 121-125. Ref.: https://goo.gl/DKUJvr
- Yu J, Cary JW, Bhatnagar D, Cleveland TE, Keller NP, et al. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl Environ Microbiol. 1993; 59: 3564-3571. Ref.: https://goo.gl/NMdDbf
- Chauvaux S, Chevalier F, Le Dantec C, Fayolle F, Miras I, et al. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J Bacteriol. 2001; 183: 6551-6557. Ref.: https://goo.gl/pE99zQ
- Lin JQ, Zhao XX, Zhi QQ, Zhao M, He ZM. Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Fungal Genet Biol. 2013; 56:78-86. Ref.: https://goo.gl/HtSYFA
- Liu SY, Lin JQ, Wu HL, Wang CC, Huang SJ, et al. Bisulfite sequencing reveals that Aspergillus flavus holds a hollow in DNA methylation. PLoS ONE. 2012; 7: e30349. Ref.: https://goo.gl/7htLt5
- Zhi QQ, Li JY, Liu QY, He ZM. A cytosine methyltransferase ortholog dmtA is involved in the sensitivity of Aspergillus flavus to environmental stresses. Fungal Biol. 2017; 121: 501-514. Ref.: https://goo.gl/wv4WPa
- Zhi QQ, Xie YY, He ZM. Genome mining for aflatoxin biosynthesis. Fungal Genom Biol. 2013; 3: 108. Ref.: https://goo.gl/cGGv39
- Zhao X, Spraker JE, Bok JW, Velk T, He ZM, Keller NP. A cellular fusion cascade regulated by LaeA is required for sclerotial development in Aspergillus flavus. Front Microbiol. 2017; 8: 1925. Ref.: https://goo.gl/N6zB77
- Zhao X, Zhi QQ, Li JY, Keller NP, He ZM. The antioxidant gallic acid inhibits aflatoxin formation in Aspergillus flavus by modulating transcription factors FarB and CreA. Toxins (Basel). 2018; 10: 270. Ref.: https://goo.gl/PczwsJ
- Chang PK, Skory CD, Linz JE. Cloning of a gene associated with aflatoxin B1 biosynthesis in Aspergillus parasiticus. Curr Genet. 1992; 21: 231-233. Ref.: https://goo.gl/oTTyhx
- Lin JQ, Zhao XX, Wang CC, Xie Y, Li GH, et al. Transcriptomic profiling of Aspergillus flavus in response to 5-azacytidine. Ann Microbiol. 2012; 63: 763-769. Ref.: https://goo.gl/46h5qV
- Nierman WC, Yu J, Fedorova-Abrams ND, Losada L, Cleveland TE, et al. Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announc. 2015; 3. Ref.: https://goo.gl/7ijZGU