Research Article
Isolation and biochemical characterization of Plant Growth Promoting (PGP) bacteria colonizing the rhizosphere of Tef crop during the seedling stage

Zerihun Tsegaye*, Birhanu Gizaw, Genene Tefera, Adey Feleke, Solomon Chaniyalew, Tesfaye Alemu and Fasil Assefa
Addis Ababa University, Department of Microbial, Cellular and Molecular Biology and Ministry of Innovation and Technology, Ethiopia
*Address for Correspondence: Zerihun Tsegaye, Department of Microbial, Cellular and Molecular Biology, Ethiopian Biodiversity Institute, Ethiopia, Tel: 251+936451046; Fax: #0116613722; Email: zerihuntsegaye1970@gmail.com
Dates: Submitted: 01 February 2019; Approved: 27 March 2019; Published: 28 March 2019
How to cite this article: Tsegaye Z, Gizaw B, Tefera G, Feleke A, Chaniyalew S, et al. Isolation and biochemical characterization of Plant Growth Promoting (PGP) bacteria colonizing the rhizosphere of Tef crop during the seedling stage. J Plant Sci Phytopathol. 2019; 3: 013-027. DOI: 10.29328/journal.jpsp.1001027
Copyright License: © 2019 Tsegaye Z, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Biolog; Plant growth promoting rhizobacteria; Tef; Rhizosphere; Rhizoplane
Abstract
The use of novel PGPR as bio inoculant is an alternative sustainable agricultural practice to improve soil health, grain quality, increase crop productivity, and conserve biodiversity. The aim of this study is to isolate, and characterized PGP bacteria colonizing tef rhizosphere during the seedling stage. For this concern, 426 samples of tef (Eragrostis tef) rhizosphere soils and roots were collected from East Shewa zone, Oromia regional state. 200 morphologically different bacterial pure colonies were isolated and screened for their PGP traits and biocontrol properties. Among these 40.5% isolates were positive for phosphate solubilization. 36% were positive for IAA production, 4.5% were positive for ammonia production, 19 % were positive for (EXPS), 15.5% were positive for protease production, 12.5% were positive for HCN productions, 9.5 % were positive for cellulase production, 4% were positive for amylase production, 3.5% were positive for chitinase production. For abiotic stress tolerance test, all of the isolates were grown well at 20oc and 30oc and neutral pH, 27% isolates were grown well at 4oc, 25.5% grew at 40oc, 25.5% were grown well on pH-9 and pH-11, 23.5% were tolerated pH-5, 3.5% grew at 50oc and 60oc, 13.5% were grown well on 5% NaCl (w/v), 3.5% were grown well on 10 and 15% NaCl (w/v), which indicated these isolates can survive in some extreme conditions. Totally 15 bacterial species having PGP traits, biocontrol properties, and abiotic stress tolerance ability were identified using the Biolog bacterial identification system. Among these, the majority of the identified PGPR have utilized carbohydrate, carboxylic acid, and amino acid, which are the main components of plant root exudates. The above results indicated that thus PGPR can be used as biofertilizers as well as biocontrol agents to replace agrochemicals to improve crop productivity. Hence, these species can be further formulated and used for greenhouse and field applications.
Introduction
Tef (Eragrostis tef) is an indigenous cereal crop of Ethiopia and it has been cultivated for thousands of years in Ethiopian high lands. It is a daily staple food for about 50 million inhabitants accounting for 14% of all calories consumed [1]. It is used to make injera, a delicious traditional fermented pancake. Porridge, local drinks like ‘tella’ and ‘katikalla’ can be prepared by using tef products as an ingredient in the process. Its straw is highly valued and used as feed for animals [2] in addition; the straw is incorporated with mud to reinforce and used for plastering walls of houses. Nutritionally tef’s grain contains14-15% protein, 72.1-75.2% complex carbohydrate, and 2% fat [1]. For human health, the benefit of tef is the high fiber content of the grain. This is predominantly important in dealing with diabetes (low glycemic index) and gluten-free, preventing anemia. It is an excellent source of essential amino acids, especially lysine. Lysine is the amino acid that is most often deficient in other grain foods [3]. It has also contained calcium, potassium and other essential minerals found in an equal amount of other grains. Tef adapted to a wide range of environments and cultivated under diverse agro-climatic conditions [4]. It performs well in both waterlogged Vertisols and clay soils in the highlands as well as water-stressed areas in the semi-arid regions throughout the country [5]. It can be grown from sea level up to 2800 meters above sea level under various rainfalls, temperatures, and soil types.
The annual area covered by tef during the main season is about 1.91 million ha, that is, accounted for 29% of total cereal area in Ethiopia which stands βirst in its area coverage [6]. Tef is, however, a very delicate and fragile crop that requires a lot of work and care, and it has one of the lowest yields of the cereal crops grown in the county. Its yield reduction is mainly due to soil infertility, especially nitrogen and phosphorus deβiciencies and logging problem (Seyifu, 1993). Like most other crops tef production is also limited by factors like disease, weed, and insects. Though tef is relatively less affected by plant diseases compared with other cereal crops, in Ethiopia, in areas where humidity is high, rusts and head smuts are important diseases of tef. Bekele [7], reported that in Ethiopia there are 22 fungi and 3 pathogenic nematodes associated with tef.
Nowadays higher grain yield of tef was recorded by applying agrochemicals such as chemical fertilizer, herbicide, and pesticide. It is an important chemical used to inhibit weeds, fungal pathogens, improve soil nutrients and stimulate plant growth. Continuous use of these agro-chemicals affects the environment, animal and human health, and biodiversity of rhizosphere in the long run. Recent research reports showed that increased use of chemical fertilizers causes groundwater contamination, which in turn causes gastric cancer, goiter, birth malformations, hypertension, testicular cancer and stomach cancer [8]. However, chemical fertilizers are neither easily available nor affordable for the majority of poor Ethiopian farmers. The use of PGP rhizobacteria as bio inoculant is an alternative sustainable agricultural practice to improve soil health and increase crop productivity, grain quality, and also conserve biodiversity [8].
Rhizobacteria are rhizosphere-competent bacteria that aggressively inhabiting plant roots; they are able to multiply and colonize all the ecological niches found on the roots at all stages of plant growth, in the presence of a competing micro-flora [9]. Its diversity in agroecosystems may be affected by soil type, agro-climatic conditions, plant species, plant-microorganism interaction, land use, and management. Each plant species has a significant effect on the rhizosphere bacterial community structure due to the differences in root exudation [10] and may select own specific microbial populations in its rhizosphere.
Rhizobacteria exert the beneficial effects on the growth of the host plant through various mechanisms are termed as PGPR [11]. It grows in, on, or around root plant tissues that stimulate plant growth, protects plants from plant pathogens and abiotic stress condition [12].
According to published reports and hypothesis, the PGPR mainly affect plant growth through several mechanisms: (i) synthesis of phytohormones, which can be absorbed by plants, (ii) fix atmospheric nitrogen (iii) mobilization of soil compounds, making them available for the plant to be used as nutrients, (iv) protection of plants under stressful conditions, thereby counteracting the negative impacts of stress, or (v) defense against plant pathogens, reducing plant diseases or death. Any bacterium possessing one or more of these above characters are known as PGPR. Several PGPR has been used worldwide for many years as bioinoculants to improve crop yields, quality, and soil fertility and contribute to more sustainable agriculture.
The success and efficiency of PGPR as bioinoculants are influenced by various factors, among which the ability of these bacteria to colonize plant roots, the exudation by plant roots and the soil health. The root colonization efficiency of PGPR is closely associated with microbial competition and survival in the soil, as well as with the modulation of the expression of several genes and cell to cell communication via quorum sensing [13]. Plant roots react to different environmental conditions through the secretion of a wide range of compounds which interfere with the plant-bacteria interaction, being considered an important factor in the efficiency of the inoculants [14]. The soil is another significant factor that affects the inoculation efficiency, due to several characteristics such as soil type, nutrient pool, soil moisture, and soil disturbances caused by management practices. The primary aim of this study is to isolate, biochemically characterize and identify plant growth promoting rhizobacteria colonizing the rhizosphere of tef crop during the seedling stage to prepare bio inoculants which are environmentally friends and economically wise to maintain sustainable crop productivity and production without harming the environment, human and animal health.
Methodology
Study time and area
This study was conducted in East Shewa Zone, Oromia Regional State, Ethiopia from 2009 to 2010 E.C. East Shewa zone has 10 districts. According to zonal statistics and information center, the zone is found between 38o57’and 39o32’ E and 7o12’ and 9o14’N and classified into three traditional agro-climatic zones. These traditional classes include 37 % Kolla (lowlands between 500 and 1,500 meters), 61% Woina Dega (midlands between 1,500 and 2,300 meters), 2% Dega (highlands between 2,300 and 3,200 meters) [15]. The Ethiopian traditional climatic zone classification system uses altitude and mean daily temperature to divide the country into 5 zones [16]. East Shewa was selected for this study because the topography of the zone land is characterized by diverse geomorphologic features distributed over the three major traditional agro-climatic zones, variability in their soil types, and rainfall. The average altitude is 1600 m but rises up to 2420 m at the northwestern and it falls to 900-1000 m towards the northeast. East Shewa zone represents Ethiopia in terms of the above-listed factors (Figure 1).
| No | Sample type | Kolla | Woina Dega | Dega | Total samples |
| 1 | Tef rhizosphere soil | 78 | 129 | 6 | 213 |
| 2 | The root of cultivated tef crops | 78 | 129 | 6 | 213 |
| Total | 156 | 258 | 12 | 426 |
Sample size and type
A total of 426 samples (213 rhizosphere soil and 213 roots) of cultivated tef varieties were collected along the traditional agro-climatic zone percentile composition during the seedling stage.
Sample collection methods
During the sampling period, six potential tef growing districts were selected according to their traditional agro-climatic zone percentile composition, Woina Dega (Ada’a, Lume, Adama, and Dugdabora), Kolla (Boset and Adama) and Dega (Gimbichu).
Each selected districts were clustered based on their soil type variation. Within each soil type tef, farmlands were clustered based on tef variety. Then within each clustered representative farmland, sampling was selected randomly. At each sampling site, triplicates of tef crop roots were uprooted with adhering soil from three different sites (1stcorner, center, and last corner) and placed into sterile test tubs to make composite root samples.
Rhizosphere soil (approximately 50g) was collected at a depth of 0cm, 5cm and 15cm from each sampling sites and placed into a sterile plastic bag to make soil composite samples. A total of 426 samples of cultivated tef rhizosphere soil and root with adhering soils were collected and kept in ice-box and transported to EBI microbiology laboratory. All soil and root samples were preserved at 4ºC until use.
Bacterial isolation
For isolation of rhizosphere bacteria, 1.0 gram of the soil was mixed in 9.0 ml of saline solution (0.9% NaCl) (w/v). The soil suspension was vortexed for five minutes to remove soil, stones, debris and dead bacterial cells. For isolation of root adhering (rhizoplane) bacteria fresh roots were washed using distilled water. For isolation of endophytic bacteria, roots were surface sterilized in70% ethanol for three minutes, followed by2% sodium hypo chlorate washing for 2 minutes and rinsing with sterile water. Roots were homogenized with a sterile mortar and pestle. Homogenized root samples were added into test tubs containing 30ml of distilled water for the purpose of making solution. Soil suspension, root washing solutions, and homogenized roots solution were serially diluted (10-1to 10-6) respectively. 100μl of supernatant from each dilution of rhizosphere soil, root washing solution and root endophytic solutions were transferred onto different prepared medium and incubated at the appropriate temperature. The purified bacteria cultures were identified based on their morphology (colonial and cellular) following Berge’s Manual of Determinative Bacteriology (Berge, 1984) and screened for different PGP traits, biocontrol properties, and abiotic stress tolerance activities.
In vitro screening of bacterial isolates for their plant growth promoting traits
Test for phosphate solubilization activity: Bacterial isolates were screened in vitro for their phosphate solubilizing activity using Pikovaskaya’s medium. The cultures were spot-inoculated on the Pikovskay’s medium plates and incubated at 30°C for 7 days. The appearances of the clear zone around bacterial growth were indicated a positive result for phosphate solubilization [17].
Test for organic acid production: The bacterial organic acid production test was conducted by inoculating test bacterial culture in minimal salt medium (MM9 broth) for 2 to 3days at 30oC. The appearance of pink color in the medium indicates organic acid formation using methyl red as an indicator [18].
Production of indole acetic acid (IAA): IAA production was detected as described by Brick et al. [19]. Bacterial cultures were grown on Luria Bertani (LB) broth amended with 100mg/l tryptophan as the precursor of IAA and incubated in a shaker at 250 rpm at 30oc for 3 to 5days. Fully grown cultures were centrifuged at 10,000 rpm for 10 min. 2ml of the supernatant was mixed with 4ml of the salkowski reagent (50 ml, 35% of HClo4, 1ml 0.5 m FeCl3 solution). Appearances of pink color in test tubes were indicating a positive result for IAA production.
Production of ammonia: Bacterial isolates were screened for the production of ammonia in peptone water described by Ajay Kumar et al. (2012). Freshly grown cultures were inoculated in 10ml peptone water containing tubs and incubated for 48 h at 30oC. Nessler’s reagent (0.5 ml) was added in each tube. Formation of brown to yellow color indicates a positive result for ammonia production.
In vitro screening of bacterial isolates for their biocontrol properties
Screen for hydrogen cyanide (HCN) production: Bacterial isolates were screened for HCN production by the methodology described by Castric [20]. The isolates were inoculated on the nutrient media plates containing 4.4 g glycine per liter. To the top of the plate, Whatman filter paper no. 1 soaked in 2% sodium carbonate in 0.5% picric acid solution was placed and sealed with parafilm. The plates were incubated at 30°C for 4 days and observed for a color change of the filter paper from deep yellow to reddish-brown indicated production of HCN [21].
Screening bacterial isolates for hydrolytic enzyme production: Bacterial isolates were screened for their hydrolytic enzyme production like protease, cellulase, amylase, and chitinase.
Protease production activity: Bacterial isolates were screened for their ability to produce proteolytic enzymes onto skim milk agar or SMA (3% v/v) medium [22]. The diameter of the clear zone formed around the bacterial colonies was measured after 48 h of incubation at 30°C.
Cellulase production activity: bacterial isolates were performed for cellulase production activity by spot inoculation on the cellulose agar media with the following composition [23], KH2PO4 0.5 g, MgSO4 0.25 g, cellulose 2 g, agar 15 g, and gelatin 2 g; distilled water one liter and at pH 6.8–7.2. The use of Congo-Red as an indicator of cellulose degradation in an agar medium provides the basis for a rapid and sensitive screening test for cellulolytic bacteria. Bacterial isolates showing clear halo zone on cellulose medium was indicated a positive result for cellulose synthesis.
Production of amylase (starch hydrolysis): The bacterial isolates were spot inoculated on starch agar (Beef extract 3.0, peptone 5.0, soluble starch 2.0, Agar 15.0, Distilled water 1liter) medium plates and incubated at 30°C for 48 h. At the end of the incubation period, the plates were flooded with iodine solution, kept for a minute and then poured off. Iodine reacts with starch to form a blue color compound. This blue color fades rapidly. Hence the colorless zone surrounding colonies indicates the production of amylase [24].
Production of chitinase: The qualitative assay for chitin production of the bacterial isolates was performed by chemical extraction of colloidal chitin from snail shell and used component medium (Colloidal chitin 4.0, K2HPO4 0.7, KH2PO4 0.3, MgSO40.5, FeSO4 0.01, ZnSO4 0.001, MnCl2 0.001, Agar 15.0, Distilled water 1000ml) for 5 to7 days at 30oC. Development of Clearance halo zone around the growing colony after the addition of iodine was considered as positive for chitinase enzyme production [25].
Biochemical characterization and identification of PGP bacteria
Phenotypic identification of the bacterial isolates was conducted using biochemical tests by the procedures described in the Bergey’s Manual of Systematic Bacteriology. Preliminary identification of PGPR isolates was performed using morphological characterization. After 24 h of growth on peptone Agar at 30°C, colonies of purified bacterial isolates were characterized for their following traits: color, shape, length, breadth and width, surface, opacity, and texture. Cellular morphology, size and division mode was detected by using light microscopy.
Carbon source utilization (biochemical) patterns of bacterial isolates having plant growth promoting traits were analyzed using Biolog OmniLog® ID System which is now widely available to assess functional diversity of microorganisms in the rhizosphere. Biolog is patented technology and automated/semi-automated biochemical tests that analyze microbial ability to utilize particular carbon sources, and chemical sensitivity assays. The Biolog OmniLog Identification system is that characterizes the ability of microorganisms to utilize or oxidize (or simply “Biolog” Inc, Hayward, California), a system that used Gene III microplate containing 94 phenotypic a panel of 71 carbon sources assays and 23 chemical sensitivity assays including pH and salt. The test panel provides a “Phenotypic fingerprint” of the microorganism that can be used to identify it at the species and strain level. The plates contained 96 wells, with a dehydrated panel of necessary nutrient medium and tetrazolium violet. Tetrazolium violet is incorporated in each of the substrates contained in a 96 well of the microplate. As a bacterium begins to use the carbon sources in certain wells of the microplate, it respires. With bacteria, this respiration process reduces a tetrazolium redox dye and those wells change color to purple. The end result is a pattern of colored wells on the microplate that is characteristic of that bacterial species.
PGPR bacterial isolates were prepared according to the manufacturer’s instructions in the OmniLog ID System User Guide (Biolog, Hayward, CA). All isolates were transferred on a Biolog Universal Growth (BUG) agar and incubated at 30°C for 24 hours. After an incubation period, Colonies were picked with a sterile wooden Biolog Streakerz™ stick and rubbed around the walls of a test tube containing inoculating fluid (A) protocol and measure absorbance/optical density (from 90 to 98) of the inoculum using Biolog turbidimeter. 100 μL of the bacterial suspension was transferred into 96 wells of GenIII microplates using multi-channel pipettor and incubated at 30°C. Read the microplate using Biolog’s microbial identification systems software (OmniLog® Data Collection) after the period of 18, 24 and 48 hours incubation. Each metabolic profile of inoculated PGPR was compared with the Omnilog Biolog database (Biolog, Hayward, CA) and identified at species and strain level [26].
Methods of Data Analysis
Data analysis was carried out using chart, table, frequency, and percentiles to evaluate PGP traits, biocontrol properties, and abiotic stress tolerance capacity of cultivable potential plant growth promoting rhizobacterial isolates.
Results
Bacterial isolation
For the preliminary morphological characterizations of potential PGPR, 200 pure bacterial colonies colonizing tef rhizosphere during seedling stage were isolated and characterized for their colonial traits such as size, shape, color, margin, elevation, and opacity. All isolates were characterized for cellular morphology using light microscopy. The highest percentage occurrence of the tef rhizosphere bacterial isolates on culture media 84.5% (169) were Gram-negative and 15.5 % (31) were Gram-positive, and among bacterial genus 56 % (112) Pseudomonas,12.5 % (25) Bacillus, 8.5 % (17) Enterobacter,7.5 % (15) Serratia, 6.5 % (13) Chryseobacter, 5 % (10) Citrobacter, 3 % (6) Flavobacter and 1 % (2) Klebsiella (Table 1) (Figure 2).
| Table 1: Morphological characterization of rhizobacterialisolates. | |||||||||
| Code | Percent ofoccurrence | Morphologicalcharacterization | |||||||
| Colonialcharacterization | Cellularcharacterization | ||||||||
| 1st field | No | Occurance (OC)% | size | shape | color | margin | Elevation | Opacity | Gram staining +/- |
| soil 1 | 18 | 9 | medium | circular | brown | entire | raised | Translucent | - |
| soil 5 | 25 | 12.5 | large | irregular | brown | curled | flat | Shiny | - |
| soil 13 | 17 | 8.5 | large | irregular | white | mucoid | raised | Transparent | - |
| soil 19 | 21 | 10.5 | medium | circular | white | entire | raised | Translucent | - |
| soil 60 | 23 | 11.5 | medium | oval | white | entire | raised | Translucent | - |
| soil 61 | 8 | 4 | medium | circular | white | entire | raised | Translucent | + |
| soil 65 | 10 | 5 | medium | circular | white | entire | raised | Transparent | - |
| soil 68 | 15 | 7.5 | medium | circular | red | entire | raised | Opaque | - |
| RW 11 | 12 | 6 | medium | circular | brown | entire | raised | Translucent | - |
| RW 22 | 6 | 3 | medium | oval | white | entire | raised | Transparent | - |
| RW29 | 3 | 1.5 | medium | circular | brown | entire | raised | Translucent | - |
| RW 30 | 2 | 1 | medium | circular | white | entire | raised | Transparent | - |
| RW 35 | 10 | 5 | medium | circular | white | entire | raised | Translucent | - |
| RW 36 | 13 | 6.5 | medium | circular | yellow | entire | raised | Translucent | - |
| RW 37 | 17 | 8.5 | medium | circular | brown | entire | raised | Translucent | + |
| -= Gram-negative, + = Gram-positive. | |||||||||
Bacterial isolates screened for plant growth promoting traits
200 pure bacterial isolates were screened for different PGP traits such as phosphate solubilization, organic acid production, IAA production, and NH3production, among these, 40.5% (81) bacterial isolates were found positive for phosphate solubilization, 35.5% (68) for IAA production, 15.5% (31) for organic acid production and 4.5% (9) for ammonia production (Table 2).
| Table 2: Bacterial isolates screened for plant growth promoting traits. | |||||||||
| Cod | No. of isolates | P-solubilization | Organic acid production | IAA production | NH3 production | ||||
| clearance | %OC | clearance | %OC | clearance | %OC | clearance | %OC | ||
| soil 1 | 7 | ++ | 3.5 | +++ | 3.5 | +++ | 3.5 | - | - |
| soil 5 | 10 | ++ | 5 | +++ | 5 | +++ | 5 | --- | - |
| soil 13 | 7 | +++ | 3.5 | ++ | 3.5 | +++ | 3.5 | +++ | 3.5 |
| soil 19 | 8 | + | 4 | - | - | - | - | - | - |
| soil 60 | 12 | + | 6 | - | - | +++ | 6 | - | - |
| soil 61 | 3 | +++ | 1.5 | - | - | ++ | 1.5 | --- | - |
| soil 65 | 4 | +++ | 2 | - | - | +++ | 2 | --- | - |
| soil 68 | 6 | +++ | 3 | - | - | ++ | 3 | - | - |
| RW 11 | 5 | ++ | 2.5 | - | - | ++ | 2.5 | - | - |
| RW 22 | 2 | + | 1 | + | 1 | +++ | 1 | -- | - |
| RW29 | 1 | +++ | 0.5 | - | - | - | - | +++ | 0.5 |
| RW 30 | 1 | ++ | 0.5 | - | - | - | - | ++ | 0.5 |
| RW 35 | 4 | +++ | 2 | - | - | +++ | 2 | - | - |
| RW 36 | 6 | +++ | 3 | - | - | +++ | 3 | - | - |
| RW 37 | 5 | +++ | 2.5 | ++ | 2.5 | ++ | 2.5 | - | - |
| % OC | 81 | 40.5% | 15.5% | 35.5% | 4.5% | ||||
| - = No production, + = weak production, ++ = medium production, +++ = high production | |||||||||
Bacterial isolates screened for bacterial biocontrol properties
200 bacterial isolates were screened for bacterial biocontrol properties such as lytic enzyme production, HCN production, and exopolysaccharide (Exp) production [27]. The percentage occurrences of the isolates were screened for bacterial biocontrol properties, 15.5 % (31) isolates were positive for protease production, 9.5 % (19) were positive for cellulase production, 7.5 % (15) were positive for amylase production, and 3.5% (7) were positive for chitin production, 12.5 % (25) were positive for HCN production, 19 % (38) were positive for Exp production (Table 3).
| Table 3: Bacterial isolates screened for bacterial biocontrol properties. | ||||||||||||
| Isolates cod | Isolates numbers | Lytic enzyme production | Production of | |||||||||
| Protease< | Cellulase | Amylase | Chitin | HCN | EXP | |||||||
| Clear | OC% | Clear | OC% | Clear | OC% | OC% | Pink | OC% | Precipitate on | OC% | ||
| soil 1 | 7 | - | 0 | ++ | 3.5 | - | 0 | 3.5 (+++) | +++ | 3.5 | - | |
| soil 5 | 10 | +++ | 5 | - | 0 | 0 | - | + | 5 | - | ||
| soil 13 | 7 | -- | 0 | __ | 0 | ++ | 2.5 | _ | _ | +++ | 3.5 | |
| soil 19 | 8 | -- | 0 | _ | 0 | _ | 0 | _ | _ | +++ | 4 | |
| soil 60 | 12 | - | 0 | _ | 0 | _ | 0 | _ | _ | _ | ||
| soil 61 | 3 | - | 0 | _ | 0 | _ | 0 | _ | _ | - | ||
| soil 65 | 4 | +++ | 2 | _ | 0 | _ | 0 | _ | _ | + | 2 | |
| soil 68 | 6 | -- | 0 | - | 0 | - | 0 | _ | +++ | 3 | + | 3 |
| RW 11 | 5 | +++ | 2.5 | ++ | 2.5 | + | 2.5 | _ | - | - | ||
| RW 22 | 2 | --- | 0 | ++ | 1 | + | 1 | _ | +++ | 1 | + | 1 |
| RW29 | 1 | +++ | 0.5 | - | 0 | - | 0 | _ | - | - | ||
| RW 30 | 1 | -- | 0 | - | 0 | + | 0.5 | _ | - | +++ | 0.5 | |
| RW 35 | 4 | - | 0 | - | 0 | - | 0 | _ | - | +++ | 2 | |
| RW 36 | 6 | +++ | 3 | - | 0 | - | 0 | _ | - | ++ | 3 | |
| RW 37 | 5 | +++ | 2.5 | +++ | 2.5 | - | 0 | _ | - | - | - | |
| % OC | 40.5 | 15.5 | 9.5 | 7.5 | 3.5 | 12.5 | 19 | |||||
| - = No production, + = weak production, ++ = medium production, +++ = high production | ||||||||||||
Bacterial isolates screened for abiotic stress tolerance activities
All bacterial isolates were screened for abiotic stress tolerance activities such as different salt concentration, different pH ranges, and different temperature ranges (Table 4). Among these 13.5 % (27) bacterial isolates were tolerated 5% salinity, 3.5 % (7) were tolerated 10 % salinity, 3.5 % (7) were tolerated 15 % salinity. For pH tolerance test, all bacterial isolates grew well on pH-7, 25.5 % (51) grew well on pH-9 and pH-11, 23.5 % (47) grew well on pH-5, and there is no growth on pH- 4 and pH- 13. For temperature tolerance test, all bacterial isolates grew well at 20 and 30oc, 27 % (54) were grown well at 4oc, 25.5 % (51) grew well at 40oc, 3.5% (7) grew well at 50oc and 3.5 % (7) grew well at 60oc (Figure 3).
| Table 4: Bacterial isolates screened for abiotic stress tolerance activities. | |||||||||||||||||
| No | Bacterial Isolates | Abiotic stress tolerance | |||||||||||||||
| Salt. conc NaCl (w/v) | pH | Temperature | |||||||||||||||
| 5 | 10 | 15 | 20 | 4 | 5 | 7 | 9 | 11 | 13 | 4 | 20 | 30 | 40 | 50 | 60 | ||
| 1 | soil 1 | - | - | - | + | + | - | - | - | + | + | + | - | -- | -- | ||
| 2 | soil 5 | - | - | - | - | - | + | + | - | - | - | + | + | + | + | -- | -- |
| 3 | soil 13 | + | - | - | - | - | + | + | + | - | - | + | + | + | -- | -- | |
| 4 | soil 19 | - | - | - | - | - | + | + | + | - | + | + | + | - | -- | -- | |
| 5 | soil 60 | - | - | - | - | - | + | + | + | - | + | + | + | + | -- | -- | |
| 6 | soil 61 | + | - | - | - | - | - | + | + | + | - | + | + | + | + | -- | -- |
| 7 | soil 65 | + | - | - | - | - | - | + | + | + | - | + | + | + | + | -- | -- |
| 8 | soil 68 | - | - | - | - | - | + | + | + | + | - | + | + | + | + | -- | -- |
| 9 | RW 11 | + | - | - | - | - | + | + | + | + | - | + | + | + | - | -- | -- |
| 10 | RW 22 | - | - | - | - | - | - | + | + | + | - | - | + | + | - | -- | -- |
| 11 | RW29 | - | - | - | - | - | + | + | + | + | - | - | + | + | - | -- | -- |
| 12 | RW 30 | + | - | - | - | - | + | + | + | + | - | + | + | + | - | -- | -- |
| 13 | RW 35 | - | - | - | - | - | + | + | + | + | - | - | + | + | - | -- | -- |
| 14 | RW 36 | - | - | - | - | - | + | + | - | - | - | - | + | + | + | -- | -- |
| 15 | RW 37 | + | + | + | - | - | + | + | - | - | - | - | + | + | + | + | + |
| - = No growth, + = growth | |||||||||||||||||
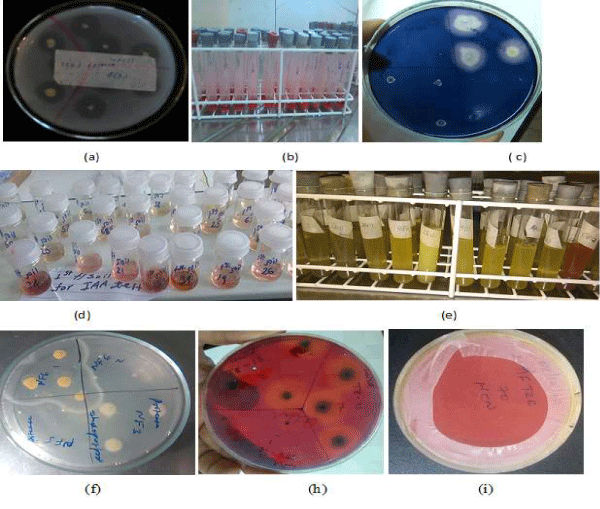
Figure 3: (a) phosphate solubilization (b)organic acid production (c)amylase production (d)IAA production (e) NH3 production (f)protease production (h)cellulase and (i) HCN production.
Biolog bacterial biochemical characterization and identification
15 tef rhizosphere bacterial species having different plant growth promoting traits, biocontrol properties, and abiotic stress tolerance ability were characterized for different Biolog carbon source utilization test. Biolog carbon sources were divided into six categories such as polymers (dextrin, β-cyclodextrin), carbohydrates (α-D-glucose, D-sorbitol, D-fructose, maltose, sucrose, arbutine, gentiobiose, and 3-methyl-glucose), carboxylic acids (pyruvic acid, lactic acid, acetic acid, citric acid, methyl pyruvate and mono-methyl-succinate), amide and amine (succinamic acid, L-alaninamide and putrescine), amino acids (D-alanine, L-alanine, L-asparagine and L-glutamic acid and L-serine), and miscellaneous (salicin, glycerol, 2,3-butanediol, 2’-deoxyadenosine, inosine, tween 80 and uridine). Among these 93.3% (14) species were utilized fructose, 73.3% (11) were utilized D-glucose, citric acid and L-alanine, 66.7% (10) were utilized mannose and lactic acid, 60 % (9) were utilized galactose and L-arginine, 53.3% (8) were utilized trehalose, 40 % (6) were utilized sucrose, 33.3% (5) were utilized raffinose and mannitol, and 20 % (3) were utilized acetic acid and pectin (Table 5)
| Table 5: Biolog bacterial biochemical characterization and identification. | ||||||||||||||||
| No | Identified PGP bacterial species | Different utilized carbon sources | Probability % | |||||||||||||
| Glucose | Fructose | Galactose | Sucrose | Mannitol | Mannose | Trahalose | Raffinose | Lactic acid | Citric acid | Acetic acid | L-alanine | L-arginine | Pectine | |||
| 1 | P. fluorescens biotype G | + | + | _ | _ | _ | + | + | _ | + | + | _ | + | + | _ | 62 |
| 2 | P. aeruginosa | + | + | - | - | - | - | - | - | + | + | - | + | + | - | 69 |
| 3 | E. cloacae ss disolvens | - | + | + | + | - | + | + | + | - | + | - | + | -- | 73 | |
| 4 | P. mendocina | + | + | - | - | - | - | - | - | + | + | + | + | + | - | 54 |
| 5 | P.putida biotype B | - | - | - | - | - | - | - | - | + | + | + | + | + | - | 56 |
| 6 | Virgibacillus sediminis | + | + | + | + | - | - | + | + | - | - | - | - | - | + | 60 |
| 7 | Citrobacter amlonaticus | + | + | + | - | - | + | - | + | + | + | - | - | - | - | 92 |
| 8 | S. marcescene ss marcescens | + | + | + | + | + | + | + | - | + | + | - | + | - | - | 89 |
| 9 | P. corrugata | + | + | + | + | + | - | - | - | + | + | - | + | + | - | 59 |
| 10 | Flavobacterium mizutai | - | + | + | + | - | + | + | + | - | - | - | - | - | + | 57 |
| 11 | P. fuscovaginae | + | + | + | - | + | + | + | + | - | + | + | - | 51 | ||
| 12 | Klebsiella oxytoca | + | + | + | + | - | + | + | + | - | - | - | + | + | - | 68 |
| 13 | P. tolaasii | + | + | + | - | + | + | + | - | + | + | + | + | + | - | 54 |
| 14 | Chryseobacterium gleum/indologenes | + | + | - | - | - | + | + | - | - | - | - | - | - | + | 97 |
| 15 | B. cereus/ pseudomycoides | - | + | - | - | + | + | - | - | + | + | - | + | + | - | 45 |
| - = No utilize carbon, + = Utilize carbon | ||||||||||||||||
Discussion
The use of novel PGP bacteria as biofertilizers, biopesticides and phytostimulator in agricultural sectors to improve crop yield, quality and maintaining the soil fertility is advisable. PGPR is the soil bacteria inhabiting around or on the root surface of plants directly involved in plant growth improvement and protects plants from disease and abiotic stresses conditions through the production of various regulatory chemicals in the vicinity of rhizosphere [28]. In this study, beneficial rhizobacteria were isolated from tef rhizosphere samples collected during the seedling stage and screened for different plant growth promoting traits, biocontrol properties and ability to tolerate abiotic stress condition. A total of 200 bacterial colonies colonizing tef rhizosphere showed diverse morphological characteristics as indicated from variations in size, shape, color, margin, elevation, and opacity. On the basis of their Gram reaction, 169 (84.5%) of the isolates were found to be Gram-negative and about 31 (15.5%) were Gram-positive.
Identification of the bacterial isolates having excellent PGP traits, biocontrol properties and abiotic stress tolerance ability using Biolog OmniLog® ID System which is now widely available to assess functional diversity of microorganisms based on carbohydrate utilization consequently, they were classified into 9 genera and 15 species such as Pseudomonas fluorescent, Pseudomonas aeruginosa, Pseudomonas mendocina, Pseudomonas putida biotype B, Pseudomonas corrugata, Pseudomonas fuscovaginae, Pseudomonas tolaasii, Enterobacter cloacae ss disolvens, Virgibacillus sediminis, Citrobacter amlonaticus, Serretia marcescen ss marcescens, Flavobacterium mizutai, Klebsiella oxytoca, Chryseobacterium gleum, and Bacillus cereus/pseudomycoide) of PGP bacteria colonizing tef rhizosphere during the seedling stage to replace agrochemicals. Hence, these species can be further formulated and used for greenhouse and field applications.
The capability of rhizobacteria to solubilize insoluble phosphates has been of interest to agricultural microbiologist as it can enhance the availability of phosphorus for the plant to improve plant growth and yield [29]. It has been reported that higher concentrations of phosphate-solubilizing bacteria are commonly found in the rhizosphere as compared to bulk soil [30]. The use of phosphate solubilizing PGPR as inoculants is one of the alternative biotechnological solutions in sustainable agriculture to meet the phosphate demands of plants. In this study, out of 200 isolates, 40.5% (81) isolates were able to solubilize phosphate at higher rates (from 20 to 25 mm) than did the other isolates. All of the above-identified PGP bacterial species found to be the most efficient phosphate solubilizes which have a great role in increasing crops productivity and production without contaminating the environment and affecting human health.
Phosphate solubilizing bacteria reduces pH of rhizosphere soils by releasing organic acids which dissolve phosphate mineral through anion exchange or chelation of Fe and Al ions associated with phosphate [31]. This process increases the availability of phosphorus for plant uptake. In this study, 15.5% isolates (Pseudomonas fluorescent biotype G, Pseudomonas aeruginosa, Enterobacter cloacae ss disolvens, Flavobacterium mizutai, Klebsiella oxytoca and Bacillus cereus/ pseudomycoide) PGPR identified from tef rhizosphere were positive for organic acid production and enhance crop yield and grain quality. The results were supported by Bharucha et al. (2013), who reported that PGPR isolated from Alfa Alfa rhizosphere soils produce organic acids and enhance plant growth.
IAA is one of the most important phytohormones which may function as an important signal molecule in the regulation of plant growth. In the present study out of 200 bacterial isolates colonizing tef rhizosphere, among these 36% isolates (P. fluorescent, P. aeruginosa, P. putida biotype B, P. corrugata, P. tolaasii, E. Cloacae ss disolvens, V. sediminis, C. amlonaticus, S. marcescen ss marcescens, F. mizutai, C. gleum, and B. cereus/pseudomycoide) are found to be good producers for IAA (Table 2). Rhizobacterial species identified from the rhizosphere are more efficient auxin producers than isolates from the non-rhizosphere soil [32]. This is an important mechanism of plant growth promotion because IAA promotes root development and uptake of nutrients [33]. It has long been proposed that IAA act coordinate demand and acquisition of nitrogen and enhance crop yields.
Nitrogen fixation ability is an important criterion for the selection of potential PGPR. In this study, the PGP bacterial colonies such as soil 13 and RW 30 (E. cloacae ass disolvens and K. oxytoca) isolated from tef rhizosphere were grew well on N-free agar media confirmed their potential of fixing atmospheric nitrogen on such media. Our result was supported by the findings of Naher et al. [34] who characterized a few N-fixing bacteria by acetylene reduction assay (ARA). Another important trait of PGPR is the production of ammonia that indirectly influences the plant growth. 4.5% of the isolates (E. cloacae ass disolvens, P. fuscovaginae, and K. oxytoca) were able to produce ammonia and enhance plant growth. The presence of ammonia producing PGP bacteria is an indicative for ammonification process were takes place in the rhizosphere than non-rhizosphere soil.
Hydrolytic enzymes act as agents for prevention of plant diseases by causing lysis of pathogenic microbes in the close vicinity of the plant as they secrete increased level of cell wall lytic enzymes (chitinase, glucanase, lipase, and proteases) [1]. In this study, 15.5 % isolates (P. aeruginosa, C. amlonaticus, P. corrugota, P. fuscovaginae, C. gleum and B. cereus/pseudomycoide), were positive for protease production, 9.5 % isolates (P. fluorescent biotype G, P. corrugata, F. mizutai and B. cereus) were positive for cellulase production, 4 % isolates (E. cloacae ss disolvens, P. corrugata, F. mizutai, and K. oxytoca) were positive for amylase production, and 3.5% isolates (P. fluorescent) were positive for chitin production. PGPR that synthesizes one or more of these lytic enzymes has been found to have biocontrol ability against a range of plant pathogenic fungi and bacteria and enhance crop yield.
HCN production by rhizobacteria has been postulated to play an important role in the biological control of pathogens (Voisard et al., 1989). In this study, 12.5% of the bacterial isolates (P. fluorescent biotype G, P. aeruginosa, S. marcescens ss marcescens, and F. mezutai) were positive for HCN production, which acts as an inducer of plant resistance. Several factors have been reported to influence the rate of HCN production. Glycine has been found to be the direct precursor of microbial cyanide production and it has been found in root exudates. HCN secreted by P. fluorescent strain CHAO has been demonstrated to stimulate root hair formation and suppress back root rot caused by Thielaviopsis basicola in tobacco plant (Voisard et al. 1989).
Exopolysaccharides (EPS) are the active constituents of soil organic matter. The important roles exhibited by EPS are (1) Protective, (2) surface attachment, (3) biofilm formation, (4) microbial aggregation, (5) plant-microbe interaction, and (6) bioremediation (Naseem and Bano, 2014). In this experiment, 19% of the bacterial isolates (E. cloacae ss disolvens, P. mendocina, C. amlonaticus, S. marcescens ss marcescens, F. mizutai, K. oxytoca, P. tolaasii, and C. gleum) were positive for exopolysaccharide production. EPS-producing plant growth-promoting rhizobacteria can significantly enhance the volume of soil macro pores and the rhizosphere soil aggregation, resulting in increased water and fertilizer availability to inoculated plants [35].
PGP bacterial abiotic stress tolerance such as (salinity, pH and extreme temperature) is a potential bioresource for improving crop productivity. Salinity is one of the most common abiotic stress factors that adversely affect plant growth and crop productivity in the world. Our results showed that 13.5 % isolates (E. cloacae ss disolvens, V. sediminis, C. amlonaticus, P. corrugata, K. oxytocaand B. cereus) were tolerated 5% salinity, 3.5 % isolates (B. cereus) were tolerated 10 % salinity, 3.5 % isolates (B. cereus/pseudomycoide) were tolerated 15 % salinity and thus are confirmed their capability of surviving in saline environment. Soil salinity has been reported to reduce plant growth, photosynthetic capacity, protein synthesis, energy and lipid metabolism, and the total nitrogen contents [36].
Soil pH is another limiting factor for PGPR. In the present study all PGP bacterial isolates grew well on pH-7, 25.5 % isolates (E. cloacae ss disolvens, P. mendocina, P. putida biotype B, V. sediminis, C. amlonaticus, S. marcescens ss marcescens, P. corrugata, F. mizutai, P. fuscovaginae, K. oxytoca, and P. tolaasii) were grew well on pH-9 and pH-11, 23.5 % isolates (P. fluorescent biotype G, P. aeruginosa, S. marcescens ss marcescens, P. corrugata, P. fuscovaginae, K. oxytoca, P. tolaasii, C. gleum and B. cereus/pseudomycoide) grew well on pH-5. Thus identified PGPR have tolerated a wide range of pH and confirmed their ability to survive both in acidic and alkaline soil and promote plant growth.
Temperature also another limiting factor that affects PGPR, in this study all PGP bacterial isolates grew well at 20 and 30oc, 27 % of isolates (P. fluorescent, P. aeruginosa, P. mendocina, P. putida biotype B, V. sediminis, C. amlonaticus, S. marcescens ss marcescens, P. corrugata and K. oxytoca) grew well at 4oc, 25.5 % isolates (P. aeruginosa, E. cloacae ss disolvens, P. putida biotype B, V. sediminis, C. amlonaticus, S. marcescens ss marcescens, C. gleum and B. cereus/ pseudomycoide) were grew well at 40oc, 3.5% (B. cereus/pseudomycoide) grew well at 50oc and 60oc. Thus PGP bacterial species were growth at extreme temperatures and promote plant growth at semiarid and arid environments.
It was clear from the present study all 15 biochemically characterized and identified PGPR were found to be the most efficient species colonizing tef rhizosphere during seedling stage were solubilize insoluble phosphorus, produced IAA, ammonia, HCN, produced lytic enzymes, produced exopolysaccharide and tolerated abiotic stress conditions. Thus are used as inoculants or bio-fertilizers as well as biocontrol agents to replace agrochemicals for improving crop productivity and production without contaminating the environment and affecting human health. Hence, these species can be further formulated and used for greenhouse and field applications.
Conclusion and Recommendation
At a global scale, the effects of continuous use of agro chemicals for improving agricultural productivity and production can cause serious damage to the human and animal health as well as the environment. It can be concluded from the above discussion that Plant growth promoting rhizobacteria are increasingly used for agricultural productivity and production improvement. In the same context, the present study was focused on the isolation, screening, and biochemically characterization of PGP rhizobacteria inhabiting tef rhizosphere during seedling stage having excellent PGP traits such as phosphate solubilization, organic acid production, Indole acetic acid and ammonia production, biocontrol properties such as hydrogen cyanide production, lytic enzymes production, exopolysaccharide production, and abiotic stress tolerance was considered for the present study. Isolates with good plant growth promoting potentialities were characterized and the best 15 efficient isolates among them were identified. The results are promising for the design of potentially active plant growth promoting PGPR strain based formulation which would be beneficial for crop improvement and crop protection. The potential of this strain could be investigated in detail and field application. These PGPR can be further explored as potential biofertilizers for sustainable agriculture.
References
- Tsegaye Z, Assefa F, Tefera G, Alemu T, Gizaw B. Characterization and Identification of Tef (Eragrostis tef) Seed Endophytic Bacterial Species and Evaluate their Effect on Plant Growth Promotion. J Plant Pathol Microbiol. 2018; 9: 438-446. Ref.: https://goo.gl/ykMZ8j
- Ketema S. Tef [Eragrostis tef (Zucc.) Trotter]. Promoting the Conservation and Use of Underutilized and Neglected Crops 12. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, Italy. 1997; 52. Ref.: https://goo.gl/5UMGqZ
- Jansen GR, DiMaio LR, Hause NL. Amino acid composition and lysine supplementation of tef. J Agric Food Chem. 1962; 10: 62-64. Ref.: https://goo.gl/rRfbFH
- Ketema S. Tef (Eragrostis tef): breeding, genetic resource, agronomy utilization and role in Ethiopia agriculture. Institute of Agricultural Research, Addis Ababa. 1993; 147. Ref.: https://goo.gl/24rvxV
- Zeleke G, Agegnehu G, Abera D, Rashid S. Fertilizer and soil fertility potential in Ethiopia: Constraints and opportunities for enhancing the system. IFPRI, Addis Ababa Ethiopia. 2010; 63. Ref.: https://goo.gl/DLkph3
- CSA (Central Statistical Agency). Agricultural Sample Survey 2008-2013 Vol. 1. Report on area and production for major crops (private peasant holding meher season) statistical Bulletin. Addis Ababa, Ethiopia. 2008.
- Bekele, E. A review of research on diseases of barley, tef and wheat in Ethiopia. 79- 08. In: Abate T. (ed.) A review of crop protection research in Ethiopia. Proceedings of the first Ethiopian crop protection symposium, 4-7 February. 1985, Addis Ababa, Ethiopia IAR, Addis Ababa, Ethiopia. 1985. Ref.: https://goo.gl/kwd62A
- Vandana UM, Chopra A, Choudhury A, Adapa D, Mazumder PB. Genetic diversity and antagonistic activity of plant growth promoting bacteria, isolated from tea-rhizosphere: a culture dependent study. Biomedical Res. 2018; 29: 853-864. Ref.: https://goo.gl/52qUST
- Muleta D, Assefa F, Granhall U. In vitro antagonism of rhizobacteria isolated from Coffea Arabica L. against emerging fungal coffee pathogens. Eng Life Sci. 2007; 7: 1-11. Ref.: https://goo.gl/drhikn
- Farina R, Beneduzia A, Ambrosinia A, de Camposa SB, Lisboab BB, et al. Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl Soil Ecol. 2012; 55: 44–52. Ref.: https://goo.gl/wScGyG
- Husen E. Screening of soil bacteria for plant growth promoting activities in vitro. J Agric Sci. 2005; 4: 27–31. Ref.: https://goo.gl/kZKznn
- Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant-soil. 2003; 255: 571–586. Ref.: https://goo.gl/CCCajt
- Alquéres S, Meneses C. Rouws L. Rothballer M. Baldani I, et al. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol Plant Microbe Interact. 2013; 26: 937-945. Ref.: https://goo.gl/1bg9bL
- Carvalhais LC, Dennis PG, Fan B, Fedoseyenko D, Kierul K, et al. Linking plant nutritional status to plant-microbe interactions. PLoS One. 2013; 8: e68555. Ref.: https://goo.gl/jL9gjP
- Gorfu D, Ahmed E. Crops and Agro-ecological Zones of Ethiopia. Ethiopian Institute of Agricultural Research. Unpublished Document. 1996. Ref.: https://goo.gl/FbKCAq
- Gemechu D. Aspects of climate and water budget in Ethiopia. Addis Ababa University Press. 1977; 76. Ref.: https://goo.gl/adVn7s
- Pikovskaya RE. Mobilization of phosphorus in soil in concentration with vital activity of some microbial species. Microbiologiya. 1948; 17: 362-370. Ref.: https://goo.gl/C2C3v3
- Trivedi MK, Patil S, Tallapragada RM. Effect of biofield treatment on the physical and thermal characteristics of vanadium pentoxide powders. J Mater Sci Eng. 2013; 11: 001. Ref.: https://goo.gl/kdvT5M
- Brick JM, Bostock RM, Silverstone SE. Rapid in situ assay for indole acetic acid production by bacteria immobilized on the nitrocellulose membrane, Appl Environ Microbial. 2004; 57: 535-538. Ref.: https://goo.gl/N2bsdL
- Castric PA. Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Can J Microbiol. 1975; 21: 613-618. Ref.: https://goo.gl/8iCuGv
- Bakker AW, Schippers B. Microbial cyanides production in the rhizosphere inrela-tion to potato yield reduction and Pseudomona sspp. Mediated plant growth stimulation. Soil Microbiol Biochem. 1987; 19: 451–457. Ref.: https://goo.gl/gta2ur
- Chang WT, Hsieh CH, Hsieh HS, Chen C. Conversion of crude chitosan to an anti-fungal protease by Bacillus cereus. World Journal of Microbiology and Biotechnology. 2009; 25: 375–382. Ref.: https://goo.gl/rru6s7
- Khianngam S, Pootaengon Y, Techakriengkrai T, Tanasupawat S. Screening and identification of cellulase-producing bacteria isolated from oil palm meal. J App Pharm Sci. 2014; 4: 090-096. Ref.: https://goo.gl/nwucT6
- Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial α-amylases: a iotechnological perspective. Process Biochem. 2003; 38: 1599-1616. Ref.: https://goo.gl/1KNB8u
- Roberts WK, Selitrennikoff CP. Plant and bacterial chitinases differ in antifungal activity. J Gen Microbiol. 1988; 134: 169-176. Ref.: https://goo.gl/Pkx4VF
- Morgan MC, Boyette M, Goforth C, Sperry KV, Greene SR. Comparison of the Biolog OmniLog Identification System and 16S ribosomal RNA gene sequencing for accuracy in identification of atypical bacteria of clinical origin. J Microbiol Methods. 2009; 79: 336-343. Ref.: https://goo.gl/KiSh3j
- Naseem H, Bano A. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. Journal of Plant Interactions. 2014; 9: 689-701. Ref.: https://goo.gl/9Ehp4X
- Tsegaye Z, Assefa F, Beyene D. Properties and application of plant growth promoting rhizobacteria. Int J Curr Trend Pharmacobiol Med Sci. 2017; 2: 30-43. Ref.: https://goo.gl/fA4aFd
- Liu M, Liu X, Cheng BS, Ma XL, Lyu XT, et al. Selection and evaluation of phosphate-solubilizing bacteria from grapevine rhizospheres for use as biofertilizers. Spanish Journal of Agricultural Research. 2016; 14: 6144-9714. Ref.: https://goo.gl/wXin4C
- Dasgupta D, Sengupta C, Paul G. Screening and identification of best three phosphate solubilizing and IAA producing PGPR inhabiting the rhizosphere of Sesbania bispinosa. International Journal of Innovative Research in Science, Engineering and Technology. 2015; 4: 3968-3979. Ref.: https://goo.gl/HGyJuz
- Habib SH, Kausar H, Saud HM, Ismail MR, Othman R. Molecular Characterization of Stress Tolerant Plant Growth Promoting Rhizobacteria (PGPR) for Growth Enhancement of Rice. Int J Agric Biol. 2015; 18: 184–191. Ref.: https://goo.gl/HhaKCG
- Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996; 42: 207-220. Ref.: https://goo.gl/1wk81p
- Carrillo AE, Li CY, Bashan Y. Increased acidification in the rhizosphere of cactus seedlings induced by Azospirillum brasilense. Naturwissenschaften 89. 202; 428-432. Ref.: https://goo.gl/KGpHcK
- Naher UA, Radziah O, Halimi MS, Shamsuddin ZH, Razi MI. Effect of root exuded specific sugars on biological nitrogen fixation and growth promotion in rice (Oryza sativa). Aust J Crop Sci. 2011; 5: 1210-1217. Ref.: https://goo.gl/aXzBgr
- Upadhyay SK, Singh JS, Singh DP. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere. 2011; 21: 214–222. Ref.: https://goo.gl/6z4Hp1
- Parida SK, Das AB. Salt tolerance and salinity effects on plants. Ecotoxicol Environ Safety. 2005; 60: 324–349. Ref.: https://goo.gl/6JrSCq
- Berg G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009; 84: 11–18. Ref.: https://goo.gl/d6VvWp
- Devi R, Thakur R. Screening and identification of bacteria for plant growth promoting traits from termite mound soil. Journal of Pharmacognosy and Phytochemistry. 2018; 7: 1681-1686. Ref.: https://goo.gl/fAkr13
- Zahid M, Abbasi MK, Hameed S, Rahim N. Isolation and identification of indigenous plant growth promoting rhizobacteria from the Himalayan region of Kashmiran their effect on improving growth and nutrient contents of maize (Zeamays L.). Front Microbiol. 2015; 6: 207. Ref.: https://goo.gl/7j4a7D