Research Article
Stability determination of candidate reference genes in cucumber plants subjected to stresses from Phytophthora melonis

Rui Wang, Hu Du, Qingmin Jin, Yu’e Lin, Yujuan Zhong, Chunpeng Yao, Xiaomei Xu and Tingquan Wu*
Vegetable Research Institute, Guangdong Academy of Agricultural Sciences/ Guangdong Key Laboratory for New Technology Research of Vegetables, Guangzhou, 510640, China
*Address for Correspondence: Tingquan Wu, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences/ Guangdong Key Laboratory for New Technology Research of Vegetables, Guangzhou, 510640, China, Tel: +86 20 38469301; Email: tingquanwu@sina.com
Dates: Submitted: 29 March 2019; Approved: 12 April 2019; Published: 15 April 2019
How to cite this article: Wang R, Du H, Jin Q, Lin Y, Wu T, et al. Stability determination of candidate reference genes in cucumber plants subjected to stresses from Phytophthora melonis. J Plant Sci Phytopathol. 2019; 3: 028-035. DOI: 10.29328/journal.jpsp.1001028
Copyright License: © 2019 Wang R, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Cucumber; Reference gene; P. melonis; Gene stability
Abstract
Stable reference genes are indispensable for ensuring the fidelity of determined gene expression levels. However, the expression levels of reference genes are unable to remain constant under all possible experimental conditions. Therefore, the stability determination of reference genes is necessary in an experimental system set. In the preset study, the stability of nine cucumber candidate reference genes (CsACT, CsUBQ, CsEF1α, CsCYP, CsαTU, CsCACS, CsTIP41, CsYSL8 and CsHEL) subjected to stresses from Phytophthora melonis(P. melonis) were determined using four different analysis methods, including Delta Ct, BestKeeper, NormFinder and GeNorm. The study results revealed that CsUBQ and CsCYP were the most stable genes suitable as internal control in cucumber plants under attack by P. melonis condition.
Introduction
Gene expression analysis is very important for the study of gene function. Quantitative RT-PCR (qRT-PCR) with reliable specificity and high sensitivity has been extensively used for analyzing the gene expression profiles [1,2]. However, qRT-PCR is dependable only when experimental data are normalized. Furthermore, the reliability of the reference gene is the most important for the normalization of data [3-5]. Some housekeeping genes involved in basic biological activities, such as ubiquitin (UBQ), actin (ACT) and elongation factor 1-alpha (EF1α), have been commonly used as an internal control, because these have been considered to express at constant levels, regardless of the experimental condition [6,7]. Since these genes are necessary for basic cellular lives, their expression levels are considered to be constant in various conditions. However, increasing studies have shown that the expression levels of these genes are not independent of experimental conditions [8,9], and the stability of reference genes differs when plants were in different stages of growth and development, or in different environmental conditions [2,9-13]. The most suitable reference genes should be constantly expressed in a plant, and should not be influenced by the internal factors of the plant and the external environment [4,14]. Therefore, it is very necessary to select a stable reference gene for the efficient quantification of target genes by qRT-PCR in given samples collected under different conditions. Delta-Ct [15], BestKeeper [16], NormFinder [17] and GeNorm [18], are the four presently available major statistical algorithms widely used for evaluating the stability of candidate reference genes.
Cucumber, which is one of the most important members of the Cucurbitaceae family, is widely cultivated worldwide. Studies on cucumber molecular biology has made significant progress [19-23], after cucumber genome sequencing data were released [24]. Cucumber blight caused by Phytophthora. melonis (P. melonis) is a destructive disease that leads to severe loss in yield and quality and yield losses of up to 80% in portions of affected fields. However, researches on the mechanism of the cucumber-P. melonis interaction have been rarely reported and there are only several papers published. CsLecRK6.1 was found that it was especially induced by P. melonis and P. capsici among the 25 CsLecRK genes of JSH cucumber plants [25] and Based on results of RNA-seq and qPCR, cucumber CsWRKY2, 20, 26, 35, 44 and 52 may be involved in disease resistance of JSH against Phytophthora melonisby SA and (or) JA signaling pathway(s) [26]. Therefore, it is of great value and significance to find one or more of the stable reference genes for studying the function of disease resistance genes under the infection conditions.
Materials and Methods
Plant materials, growth conditions and treatments
Cucumber plants from the B80 cultivar used throughout the study were cultivated in a growth chamber at 26-28°C with 85% relative humidity and a 10-hour light and 14-hour dark cycle. Ten-day old seedlings were used for the experiments. The underground parts of the plants were treated by zoospore suspension of P. melonis (5×103 zoospores/ml), while mixtures of roots and stems, and leaves of the seedlings were respectively collected for RNA extraction at 0, 6, 12, 24 and 48 hours after treatment, and there are Obvious symptoms appeared at 24 - 48 hours after inoculation: the constriction of stem base, and the plants begin to wilt. The phenotypes of cucumber plants CK (no inoculation) and 48h post-inoculation with P. melonis (5×103 zoospores/ml) were shown in Supplemental Figure 1. P. melonis were grown on V8 solid culture medium and zoospores were induced in V8 liquid culture medium. V8 juice is purchased from Campbell Soup Company. V8 liquid culture medium of 1 liter contains 200 ml of clarified V8 juice, 3 g of CaCO3, and about 800 ml of dH2O, Ph=5.8. V8 Solid culture medium of 1 liter Contains all the components of the liquid medium plus 16 g of agar, Ph=5.8.
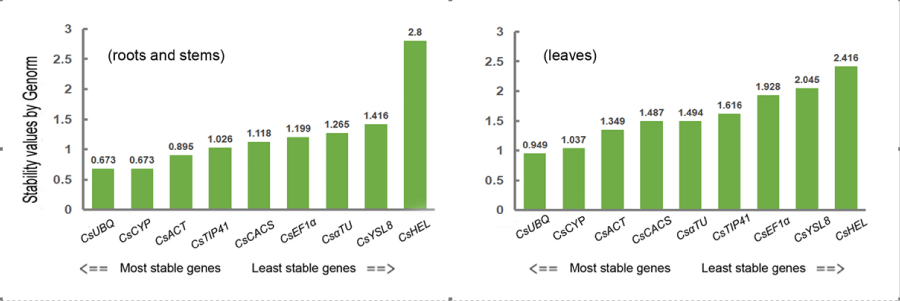
Figure 1: Ranking of the candidate reference genes according to GeNorm analysis. The values are average values of three independent biological repeat experiments. The stability value represents the stability of the gene and the most stable gene(s) has the smallest stability value(s). When the stability values of the determined genes are less than 1.5, the genes can be as the reference genes.
Experimental design
The nine cucumber genes (CsACT, CsUBQ, CsEF1s, CsCYP, CsαTU, CsCACS, CsTIP41, CsYSL8 and CsHEL) were usually chosen as the reference genes under some experimental conditions, so we predict that one or some of them may be as reference genes under the experimental condition of cucumber plants infected by P. melonis. The expression of every candidate reference gene was tested at the five time points (0, 6, 12, 24 and 48 hours) post-inoculation with P. melonis. The stability of candidate reference genes was determined using four different analysis methods, including Delta Ct, BestKeeper, NormFinder and GeNorm. Finally, we combined these results to screen out the most stable reference gene(s) in our experimental system.
RNA isolation and qPCR analysis
In order to determine the expression levels of reference genes, qRT-PCR was performed using the BIO-RAD CFX96 real-time PCR system (Bio-Rad, USA) and SYBR® Premix Ex Taq™ II mix kit (Takara). Total RNA was extracted using the RNAiso Plus reagent (takara), and first-strain cDNA was synthesized using a PrimeScript™ RT reagent Kit with the gDNA Eraser (TaKaRa Perfect Real Time), according to manufacturer’s protocol. A 10-fold dilution of the resulting cDNA was amplified employing SYBR® Premix Ex Taq™ II in a 10-µL volume with the following program: 95°C for 30 seconds, 30 cycles of 95°C for five seconds, 60°C for 15 seconds, and 72°C for 15 seconds. Three experimental and three biological replicates were carried out for assaying the expression of the reference genes.
Origin and culture of P. melonis and the preparation of zoospore suspension
P. melonis was isolated from cucumber plants infected in the experimental field of Baiyun of the Vegetable Research Institute, Guangdong Academy of Agricultural Sciences (Guangzhou), and these were cultured in V8 solid culture medium. Pieces of fresh mycelia were added in 10% V8 l culture medium for 2-3 days, the mycelia were placed in water to induce sporangia production for two days, and the water was renewed once every 12 hours. The zoospore release was induced by putting sporangia in water at 4°C for 30 minutes, the number of zoospores was determined by microscopy, and zoospore suspension were adjusted to 5×103 zoospores/ml.
Analysis method of the stability of cucumber candidate reference genes
The stability of cucumber candidate reference genes was analyzed using presently available major computational programs, such as Delta-Ct, BestKeeper, NormFinder and geNorm, in order to compare and rank the tested candidate reference genes.
Results
Transcription profiling of cucumber candidate reference genes
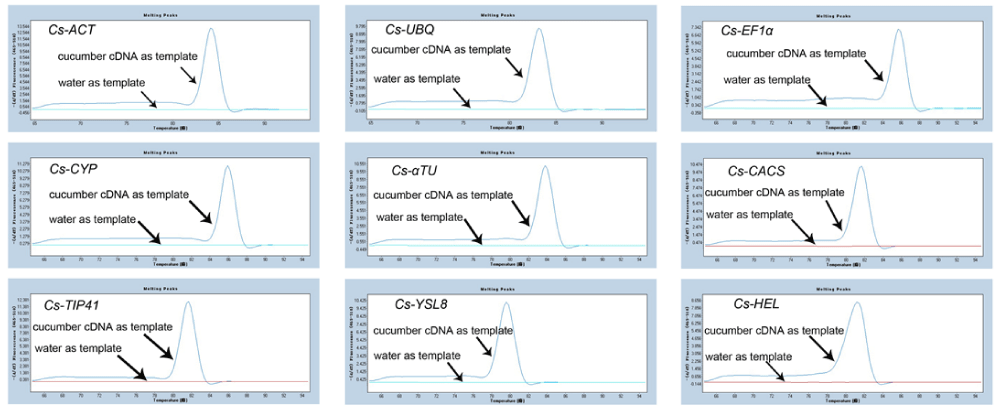
In order to identify the most stable reference genes for studies on cucumber gene expression under inoculation with P. melonis condition, the primers of nine candidate reference genes (Table 1) in cucumber were respectively designed for the qRT-PCR assay based on SYBR green detection. The specificity of the qRT-PCR amplification was confirmed through the single-peak melting curve of the PCR products (Figure 2). The expression levels of these candidate reference genes in mixture of roots and stem, and leaves were respectively presented with threshold cycle (Ct) values (Tables 2,3).
Figure 2: Melting curve analysis of the products amplified of candidate reference genes. Melting curve analysis of 9 candidate reference genes (Cs-ACT, Cs-UBQ, Cs-EF1α, Cs-CYP, Cs-αTU, Cs-CACS, Cs-TIP41, Cs-YSL8, Cs-HEL) were implemented with 0.1-0.5 uM primer final concentration and 5-50ng template cDNA final concentration. The experiments were repeated three times and the results were similar.
| Table 1 Cucumber candidate reference genes and their primer sequences and product Sizes. | |||||
| Accession number | Annotation |
Forward primer (5′-3′) | Reverse primer (5′-3′) | Product size (bp) | |
| Csa017310 | Actin (Cs-ACT) | AACTGCCCGATGGA CAAGTC |
CGGTCAGCGATACCAGGGAAC | 214 | |
| Csa000874 | Ubiquitin (Cs-UBQ) | GCGTAAGAAGAAGA CCTACACCA |
CCTTTCCAGAGTCATCGACCT | 105 | |
| Csa006172 | Elongation factor 1-alpha (Cs-EF1α) | ACTTTATCAAGAACATGATTAC | TTCCTTCACAATTTCATCG | 230 | |
| Csa014027 | Cyclophilin (Cs-CYP) | GGAAATGGTACAGGAGGTG | CATACCCTCAACGACTTGAC | 201 | |
| Csa021066 | Alpha-tubulin (Cs-αTU) | CATTCTCTCTTGGAACACACTGA | TCAAACTGGCAGTTAAAGATGAAA | 254 | |
| Csa018705 | Clathrin adaptor complex subunit (Cs-CACS) | TGGGAAGATTCTTATGAAGTGC | CTCGTCAAATTTACACATTGGT | 171 | |
| Csa003506 | PP2A phosphatase activator (Cs-TIP41) | CAACAGGTGATATTGGATTATG ATTATAC | GCCAGCTCATCCTCATATAAG | 221 | |
| Csa012471 | Mitosis protein (Cs-YSL8) | CCTTGTGGATATCACAGAAGTT | CTTGTTTATCCTTGAGTGCC | 155 | |
| Csa015849 | Helicase (Cs-HEL) | TTCTCGAAGATTTAGTGATTCAT GTG | CAATGGACGAATGCAAAGG | 168 | |
| Table 2: Ct values of cucumber candidate reference genes. | |||||||||
| Post-inoculation (h) | Cs-ACT | Cs-HEL | Cs-CYP | Cs-EF1α | Cs-TIP41 | Cs-αTU | Cs-UBQ | Cs-CACS | Cs-YSL8 |
| 0 | 21.75 | 18.39 | 20.59 | 22.41 | 26.08 | 23.96 | 21.02 | 27.01 | 29.04 |
| 6 | 21.80 | 32.01 | 19.86 | 23.08 | 26.52 | 21.66 | 20.84 | 25.68 | 28.47 |
| 12 | 20.20 | 27.72 | 18.90 | 20.27 | 24.37 | 21.11 | 19.74 | 23.95 | 27.42 |
| 24 | 21.92 | 32.44 | 19.70 | 21.06 | 25.80 | 23.26 | 20.76 | 25.97 | 27.99 |
| 48 | 22.94 | 26.85 | 20.97 | 22.96 | 26.05 | 23.69 | 21.01 | 24.84 | 27.18 |
| Notes: The samples are from roots and steam and Ct values of three repeated biological experiments. | |||||||||
| Table 3: Ct values of cucumber candidate reference genes. | |||||||||
| Post-inoculation (h) | Cs-ACT | Cs-HEL | Cs-CYP | Cs-EF1α | Cs-TIP41 | Cs-αTU | Cs-UBQ | Cs-CACS | Cs-YSL8 |
| 0 | 21.39 | 29.57 | 19.80 | 21.69 | 25.16 | 24.09 | 20.01 | 26.60 | 28.25 |
| 6 | 21.93 | 30.08 | 20.22 | 23.25 | 25.63 | 21.04 | 20.05 | 24.40 | 29.03 |
| 12 | 22.32 | 27.85 | 18.61 | 21.16 | 25.38 | 20.72 | 20.17 | 25.52 | 27.86 |
| 24 | 20.97 | 33.76 | 21.19 | 21.40 | 26.65 | 24.55 | 20.30 | 25.01 | 26.31 |
| 48 | 23.30 | 28.30 | 20.29 | 21.81 | 26.43 | 23.99 | 20.57 | 25.02 | 27.73 |
| Notes: The samples are from leaves and Ct values are average values of three repeated biological experiments. | |||||||||
Melting curve analysis of 9 candidate reference genes (Cs-ACT, Cs-UBQ, Cs-EF1α, Cs-CYP, Cs-αTU, Cs-CACS, Cs-TIP41, Cs-YSL8, Cs-HEL) were implemented with 0.1-0.5 uM primer final concentration and 5-50ng template cDNA final concentration. The experiments were repeated three times and the results were similar.
GeNorm analysis
The criteria for choosing the reference genes by GeNorm is that the most stable gene(s) has the smallest stability value(s) and when the stability values of the determined genes are less than 1.5, the genes can be as the reference genes. For cucumber roots and stems, the results revealed that except CsHEL, the stability values of other eight genes were less than 1.5 and for cucumber leaves, the stability values of five genes (CsUBQ, CsCYP, CsACT, CsCACS and CsɑTU) were less than 1.5 (Figure 2). So, CsUBQ, CsCYP, CsACT, CsCACS and CsɑTU can be as reference genes in cucumber leaves or roots and stems under the inoculation conditions of P.melonis using GeNorm analysis.
BestKeeper analysis
The criteria for choosing the reference genes by BestKeeper is that the most stable gene(s) has (or have) the smallest SD values and when SD value is less than 1, the gene can be as reference gene.
For cucumber roots and stems, BestKeeper analysis revealed that two genes (CsUBQ and CsCYP) reached the standard of the reference genes (Table 4). For cucumber leaves, BestKeeper analysis showed the same result that CsUBQ and CsCYP with SD values less than 1, reached the standard of the reference genes (Table 5). So CsUBQ and CsCYP are suitable as reference genes in cucumber roots and stems or leaves under the inoculation conditions of P.melonis using BestKeeper analysis.
| Table 4: The expression stability values of cucumber candidate reference genes in roots and stems of cucumber calculated by the BestKeeper. | |||||||||
| Experiment 1 | CsACT | CsHEL | CsCYP | CsEF1α | CsTIP41 | CsαTU | CsUBQ | CsCACs | CsYSL8 |
| geo Mean [CP] | 21.67 | 29 | 19.98 | 21.88 | 25.73 | 22.67 | 20.66 | 25.46 | 27.96 |
| AR Mean [CP] | 21.72 | 27.48 | 20.00 | 21.96 | 25.76 | 22.74 | 20.68 | 25.49 | 28.02 |
| min [CP] | 20.18 | 16.81 | 18.64 | 19.72 | 23.86 | 20.41 | 19.74 | 23.73 | 25.35 |
| max [CP] | 23.88 | 33.21 | 21.03 | 24.43 | 27.87 | 25.12 | 21.72 | 27.35 | 30.83 |
| std dev [+/- CP] | 1.24 | 4.91 | 0.73 | 1.57 | 1.16 | 1.45 | 0.60 | 1.14 | 1.52 |
| CV [% CP] | 5.68 | 19.14 | 3.68 | 7.14 | 4.47 | 6.32 | 2.84 | 4.47 | 5.44 |
| min [x-fold] | -3.24 | -32.15 | -2.63 | -4.98 | -3.70 | -4.81 | -2.23 | -3.55 | -6.31 |
| max [x-fold] | 4.7 | 26.15 | 2.15 | 6.76 | 4.62 | 5.68 | 2.16 | 3.96 | 7.34 |
| std dev [+/- x-fold] | 2.39 | 272.54 | 1.68 | 3.05 | 2.25 | 2.80 | 1.57 | 2.21 | 2.87 |
| Table 5: The expression stability values of cucumber candidate reference genes in leaves of cucumber calculated by the BestKeeper. | ||||||||||
| Experiment 1 | CsACT CsHEL | CsCYP | CsEF1α | CsTIP41 | CsαTU | CsUBQ | CsCACs | CsYSL8 | ||
| geo Mean [CP] | 21.94 | 29.76 | 19.99 | 21.80 | 25.81 | 22.79 | 20.22 | 25.67 | 27.76 | |
| AR Mean [CP] | 21.98 | 29.91 | 20.02 | 21.86 | 25.85 | 22.88 | 20.22 | 25.71 | 27.84 | |
| min [CP] | 20.29 | 26.55 | 19.00 | 19.68 | 23.96 | 20.79 | 19.74 | 23.44 | 24.76 | |
| max [CP] | 23.83 | 33.76 | 21.59 | 24.06 | 27.93 | 25.23 | 20.59 | 27.27 | 30.58 | |
| std dev [+/- CP] | 1.19 | 2.76 | 0.85 | 1.37 | 1.28 | 1.65 | 0.28 | 1.08 | 1.76 | |
| CV [% CP] | 5.43 | 9.25 | 4.23 | 6.27 | 4.92 | 7.17 | 1.37 | 4.21 | 6.27 | |
| min [x-fold] | -2.75 | -16.68 | -2.76 | -4.65 | -3.70 | -8.73 | -1.42 | -5.25 | -8.24 | |
| max [x-fold] | 3.89 | 17.92 | 3.11 | 6.01 | 4.49 | 5.54 | 1.31 | 3.12 | 7.78 | |
| std dev [+/- x-fold] | 2.32 | 6.84 | 1.80 | 2.65 | 2.47 | 3.22 | 1.22 | 2.14 | 3.52 | |
Ranking order of gene stability by Bestkeeper:
CsUBQ> CsCYP > CsCACS > CsTIP41 > CsACT > CsαTU > CsYSL8 > CsEF1α > CsHEL
(0.60) (0.73) (1.14) (1.16) (1.24) (1.45) (1.52) (1.57) (4.91)
____________________________________________________________________________________________________________________Notes: The values are average values of three independent biological repeat experiments. The most stable gene(s) has (or have) the smallest SD (std dev [+/- CP]) values and when SD values are less than 1, the genes are stability and can be as the reference genes.
Ranking order of gene stability by Bestkeeper:
CsUBQ > CsCYP > CsCACS > CsACT > CsTIP41> CsEF1α >CsαTU > CsYSL8 > CsHEL
(0.28) (0.85) (1.08) (1.19) (1.28) (1.37) (1.65) (1.76) (2.76)
______________________________________________________________________________________________________________________Notes: The values are average values of three independent biological repeat experiments. The most stable gene(s) has (or have) the smallest SD (std dev [+/- CP]) values and when SD values are less than 1, the genes are stability and can be as the reference genes.
Norm Finder analysis
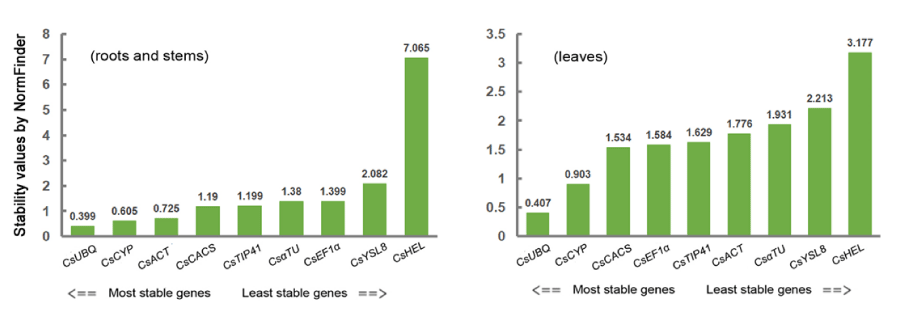
The criteria for choosing the reference genes by NormFinder is that the most stable gene(s) has (or have) the smallest stability value(s). Using the method, we can only get the ranking order of the stability of genes and choose the most stable gene as the reference gene. Nine candidate reference genes were analyzed using the NormFinder method in three repeat experiments.
For cucumber roots and stems or leaves the results revealed that CsUBQ and CsCYP ranked in the top two positions and (Figure 3). Comprehensively, CsUBQ and CsCYP are the best genes as reference genes in cucumber under P. melonis infection condition.
Figure 3: Ranking of the candidate reference genes according to NormFinder analysis. The values are average values of three independent biological repeat experiments. The stability value represents the stability of the gene and the most stable gene(s) has (or have) the smallest stability value(s).
Delta CT analysis
The criteria for choosing the reference genes by Delta CT is that the most stable gene(s) has (or have) the smallest STDEV value(s), as the similar to NormFinder. Using Delta CT method, we can get the ranking order of the stability of genes and determine the most stable gene(s). Delta CT analysis revealed that CsUBQ and CsCYP are the most stable genes in all experiments (Tables 6,7).
| Table 6: The average STDEV value and ranking order of cucumber candidate genes in roots and stems of cucumber calculated by the Delta CT | |||||||||
| CsACT | CsHEL | CsCYP | CsEF1α | CsTIP41 | CsαTU | CsUBQ | CsCACS | CsYSL8 | |
| STDEV value | 2.04 | 7.2 | 2.02 | 2.39 | 2.21 | 2.36 | 1.91 | 2.21 | 2.85 |
| Ranking order | 3 | 8 | 2 | 6 | 4 | 5 | 1 | 4 | 7 |
| Notes: The values are average values of three independent biological repeat experiments. The criteria for choosing the reference genes by Delta CT is that the most stable gene(s) has (or have) the smallest STDEV value(s) | |||||||||
| Table 7: The average STDEV value and ranking order of cucumber candidate genes in leaves of cucumber calculated by the Delta CT. | |||||||||
| CsACT | CsHEL | CsCYP | CsEF1α | CsTIP41 | CsαTU | CsUBQ | CsCACS | CsYSL8 | |
| STDEV value | 2.54 | 3.54 | 2.05 | 2.31 | 2.35 | 2.54 | 1.9 | 2.34 | 2.68 |
| Ranking order | 6 | 8 | 2 | 3 | 5 | 6 | 1 | 4 | 7 |
| Notes: The values are average values of three independent biological repeat experiments. The criteria for choosing the reference genes by Delta CT is that the most stable gene(s) has (or have) the smallest STDEV value(s). | |||||||||
Discussion
Several approaches have been proposed to evaluate the stability of reference gene expression, such as the GeNorm, BestKeeper, NormFinder and Delta-Ct method. In the present study, four analysis methods were used to compare and rank nine cucumber candidate reference genes (CsACT, CsUBQ, CsαTU, CsCYP, CsEF1α, CsCACS, CsHEL, CsYSL8 and CsTIP41) under P. melonis infection condition. We can choose genes suitable for reference genes using GeNorm and BestKeeper method, and the most stable gene(s) can be determined using NormFinder and Delta-Ct method. The analysis results are not exactly the same by these four methods, sometimes vary greatly. There are five genes (CsUBQ, CsCYP, CsACT, CsCACS and CsɑTU) suitable as reference genes by GeNorm method. And CsUBQ and CsCYP are suitable as reference genes in cucumber plants under the inoculation conditions of P.melonis using BestKeeper analysis. It’s exciting to us that CsUBQ and CsCYP are also the most stable genes using two methods of NormFinder and Delta-Ct. So CsUBQ and CsCYP are regard as the most stable genes suitable as reference genes among nine gene under the condition of cucumber plants infected by P.melonis.
In cucumber plants subjected to growth regulators and abiotic stress, CsCACS, CsTIP41 and CsEF1α are the best reference genes, while the stability rankings of CsACT, CsUBQ, CsαTU and CsCYP were ranked lower [9]. However, the study results revealed that CsUBQ and CsCYP were the most stable reference genes under the condition of cucumber plants infected by P. melonis. Our research results suggest that under the condition of biotic stress, the stability of reference genes in cucumber plants are different with those under the condition of abiotic stress or growth regulators, providing a solid foundation for future studies.
Acknowledgment
The work was supported the National Key Research and Development Program of China (2018YFD0100702), Key projects of Guangzhou science and Technology Innovation Committee (201804020047), the project “Fine Mapping of Phytophthora Blight Resistance Gene and Cloning of candidate gene(s) in South China-type cucumber” was supported by the National Natural Science Foundation of China (Grant No. 37101916), the Natural Science Foundation of Guangdong Province (2016A030313777), Special funds for agricultural industry development of Guangzhou (1710023) and the Project of “Construction of Guangdong Province Crop Germplasm Resource Bank (Nursery) and Collection, Preservation and Evaluation of Crop Germplasm Resources“.
References
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, et al. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics. 2006; 6: 47-59. Ref.: https://tinyurl.com/y4pl23bf
- Gantasala NP, Papolu PK, Thakur PK, Kamaraju D, Sreevathsa RU. Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Res Notes. 2013; 6: 312. Ref.: https://tinyurl.com/y4xoe9cz
- Thellin O, Zorzi W, Lakaye B, De-Borman B, Coumans B, et al. Husekeeping genes as internal standards: use and limits. J. Biotechnol. 1999; 75: 291-295. Ref.: https://tinyurl.com/y4cltzsy
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalization: strategies and considerations. Genes Immun. 2006; 6: 279-84. Ref.: https://tinyurl.com/y4wlytm4
- Tian C, Jiang Q, Wang F, Wang GL, Xu ZS, et al. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. Plos One. 2015; 10: e0117569. Ref.: https://tinyurl.com/y6e4nqcs
- Santi S, Schmidt W. Laser microdissection-assisted analysis of the functional fate of iron deficiency-induced root hairs in cucumber. J Exp Bot. 2008; 59:697-704. Ref.: https://tinyurl.com/y3u8yhj3
- Xia XJ, Zhang Y, Wu JX, Wang JT, Zhou YH, et al. Brassinosteroids promote metabolism of pesticides in cucumber. Agric Food Chem. 2009; 57: 8406-8413. Ref.: https://tinyurl.com/y3mxyjnd
- Robinson TL, Sutherland IA, Sutherland J. Validation of candidate bovine reference genes for use with real-time PCR. Vet Immunol Immunopathol. 2007; 115: 160-165. Ref.: https://tinyurl.com/y6hpdq7j
- Migocka M, Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Molecular Breeding. 2011; 28: 343-357. Ref.: https://tinyurl.com/y5af57ng
- Chandna R, Augustine R, Bisht NC. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS One. 2012; 7: e36918. Ref.: https://tinyurl.com/y3n85cka
- Hu R, Fan C, Li H, Zhang Q, Fu YF. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol. 2009; 10:93. Ref.: https://tinyurl.com/y2dzlz8y
- Warzybok A, Migocka M. Reliable reference genes for normalization of gene expression in cucumber grown under different nitrogen nutrition. Plos One. 2013; 8: e72887 Ref.: https://tinyurl.com/y67f7g8m
- Wan H, Zhao Z, Qian C, Sui Y, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2009; 399: 257-261. Ref.: https://tinyurl.com/y3ayy86k
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002; 29: 23-39. Ref.: https://tinyurl.com/y3nrrjmo
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006; 7: 33. Ref.: https://tinyurl.com/y6zlcchr
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-excel-based tool using pair-wise correlations. Biotechnol Lett. 2004; 26: 509-515. Ref..: https://tinyurl.com/y4y3frkb
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004; 64: 5245-5250. Ref.: https://tinyurl.com/yya9s2cj
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2012; 18: 3 (RESEARCH0034). Ref.: https://tinyurl.com/yy2kw3pu
- Song Y, Du D, Chu M, Li X, Xu H, et al. Cloning and characterization of a cucumber phospholipase D gene in response to excess nitrate and other abiotic stresses. Scientia Horticulturae. 2012; 135:128-136. Ref.: https://tinyurl.com/y3f92yns
- Wang R, Chen M, Liao F, Jiang F, Ma B, et al. Cloning of the gene encoding cucumber lumazine synthase and an analysis of its promoter activity in cucumber. Canadian Journal of Plant Science. 2010; 90: 809-818. Ref.: https://tinyurl.com/yy97ybos
- Wang M, Bi H, Liu P, Ai X. Molecular cloning and expression analysis of the gene encoding sedoheptulose-1,7-bisphosphatase from Cucumis sativus. Scientia Horticulturae. 2011; 129: 414-420. Ref.: https://tinyurl.com/y5937kcp
- Qi J, Liu X, Shen D, Miao H, Xie B, et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet. 2013; 45: 1510-1515. Ref.: https://tinyurl.com/y3hn9wys
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, et al. Development of broad virus resistance in non-transgenic cucumber using crispr/cas9 technology. Mol Plant Pathol. 2016; 17: 1140-1153. Ref.: https://tinyurl.com/y6rngnpp
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009; 475: 1275-1281. Ref.: https://tinyurl.com/y2xzsvp9
- Wu T, Wang R, Xu X, He X, Sun B, et al. Cucumis sativus L-type lectin receptor kinase (CsLecRK) gene family response to Phytophthora melonis, Phytophthora capsici and water immersion in disease resistant and susceptible cucumber cultivars. Gene. 2014; 549: 214-222. Ref.: https://tinyurl.com/yxsmlxse
- Xu X, Wang R, Chao J, Lin Y, Jin Q, et al. The expression patterns of Cucumis sativus WRKY (CsWRKY) family under the condition of inoculation with Phytophthora melonisin disease resistant and susceptible cucumber cultivars. Canadian Journal of Plant Science. 2015; 95: 1121-1131. Ref.: https://tinyurl.com/yycqkswq