More Information
Submitted: 27 April 2020 | Approved: 06 May 2020 | Published: 07 May 2020
How to cite this article: Diah Yuniti IGA, Widnyana IK, Wirawan IGP, Wijaya IN, Sritamin M. Gene polymorphisms CVPDr on some plants citrus in Bali Island. J Plant Sci Phytopathol. 2020; 4: 036-041.
DOI: 10.29328/journal.jpsp.1001049
ORCiD ID: orcid.org/0000-0002-4864-6578
Copyright License: © 2020 Diah Yuniti IGA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Citrus; CVPDr gene; Polymorphism
Gene polymorphisms CVPDr on some plants citrus in Bali Island
I Gusti Ayu Diah Yuniti1, I Ketut Widnyana1*, I Gede Putu Wirawan2, I Nyoman Wijaya2 and Made Sritamin2
1Faculty of Agriculture, University of Mahasaraswati Denpasar, Bali, Indonesia
2Faculty of Agriculture, Udayana University, Bali, Indonesia
*Address for Correspondence: I Ketut Widnyana, Agro Technology Department, Faculty of Agriculture, University of Mahasaraswati Denpasar, Bali, Indonesia, Tel: +628123950640; Email: widnyanaketut@gmail.com
Citrus Vein Phloem Degeneration (CVPD) is the main disease of citrus plants in Indonesia. This disease is caused by Gram negative bacteria, Candidatus Liberibacter asiaticus. Almost all citrus plants are susceptible to this disease and only a few citrus plants such as seedless lime (Citrus aurantiifolia var. Seedles) and kinkit citrus (Triphacia trifoliate) are tolerant. Both of these citrus plants store DNA fragments of CVPDr which are considered as tolerant factors (841 bp). However, this study found that CVPDr DNA fragments were also found in citrus plants susceptible to CVPD disease. This research aims to study DNA polymorphisms from CVPDr DNA fragments in citrus plants on the island of Bali. The PCR test showed T. trifoliate and C. aurantifolia that are resistant to CVPD and Pylogenically are in the same group as C. nobilis var Buleleng, C. reticulate var. Slayer Buleleng, and C. amblicarpa. On the other hand, citrus plants susceptible to CVPD are in a different group. There are two types of citrus plants not containing CVPDr DNA fragments, namely C. nobilis var. Petang and M. paniculata L. These results indicate that the CVPDr DNA fragment polymorphism is a factor tolerant to CVPD disease.
Citrus is one of the horticultural plants that is very important in the economy of the people of Indonesia. This plant has long been known and cultivated in Indonesia [1]. Citrus is a local fruit that is pretty much on the market, the quality of citrus fruit can be influenced by various factors including the attack of Citrus Vein Phloem Degeneration (CVPD). All cultivated citrus plants are susceptible to CVPD disease attacks [2]. On the other hand it was reported that several types of citrus plants, especially those with less economic value, were known to be resistant to CVPD. Types of citrus that are resistant to CVPD, hereinafter referred to as citrus with the CVPDr gene. Citrus plants resistant to CVPD (CVPDr) are thought to contain genes that produce a trait that is able to break the pathogenic infection of CVPD (L. asiaticum) or is able to resist transmission of pathogens carried by vector insects [3].
One gene named CVPDr gene plays a role in citrus resistance of CVPD. The CVPDr gene is found in citrus plants that are resistant or relatively resistant to CVPD diseases, namely Kinkit citrus and Seedless Lime citrus. But CVPDr gene is also found in citrus Siam Kintamani, which is a citrus crop that is susceptible to disease CVPD, so it becomes difficult to explain the mechanism of disease infection CVPD. It is possible that the CVPDr gene is not expressed so it cannot play a good role in providing resistance to CVPD disease. The CVPDr gene functions to remove obstacles in the absorption of nutrients into plant cells by the CVPD bacteria. The CVPDr gene is located on the chromosome of an citrus plant, but its molecular-level mechanism of action is unknown [3].
This study aims to determine the pathogenic CVPD in various types of citrus plants in Bali Indonesia. This study uses analysis Polymerase Chain Reaction (PCR) with O11 and O12 primers and amplified DNA fragments will be sequenced. The sequence of DNA fragments from various types of citrus plants in Bali will be analyzed to see polymorphisms that may be related to CVPD infection rates. Among all the cultivation of citrus such as Tangerines citrus, Siam citrus, Lime citrus, Lime Without Seeds citrus and other citrus fruits, are searched for the presence of genes CVPDr using PCR with primers and gene CVPDr [4].
Type of research and sample sources
This research is an experimental study conducted at the Biotechnology Laboratory, University of Udayana, Bali-Indonesia. Determination of the sample in this study using samples of citrus leaves that are resistant and susceptible to CVPD disease from several types of citrus plants namely kinkit (Triphacia trifoliate), seedless lime (C. aurantiifoliavar. Seedless), siam citrus (C. nobilis), tangerines (C. reticulate) and limes (C. amblycarpa) taken at the center of citrus plantations in Bali.
DNA isolation and DNA analysis by PCR
Total DNA from citrus plants was isolated using the NucleoSpin® Plant II kit from Marchery-Nagel. Samples were taken from leaf bones by observing the symptoms of CVPD attacks and detection using PCR analysis. Detection of the presence of the gene CVPDr performed by PCR analysis using WR-F and WR-R and use the program denaturation at a temperature of 94 ºC for 60 seconds, annealing at a temperature of 60 °C for 30 seconds, and elongation at a temperature of 72 ºC for 90 seconds. All of these stages were carried out with 36 repetition cycles [3].
Agarose gel electrophoresis and PCR visualization
Agarose gel consists of 1% agarose dissolved in 100 ml TAE buffer (consisting of 40 mM tris acetate pH 7.9; 2 mM Sodium EDTA). DNA samples (8 µl DNA + 2 µl loading dye) each were filled in the gel pit. Electrophoresis is performed with a voltage of 100 volts for ± 20 minutes. Soak in EtBr solution for ± 15 minutes. Then the results of the electrophoresis were visualized with a UV transilluminator to see the position of the bands DNA of each sample and then documented.
CPVDr gene expression by DNA sequencing analysis
Sequencing analysis Modern automated DNA sequencing machines are able to sort 384 fluorescent-labelled samples at once in a single batch (electrophoresis) that can be carried out up to 24 times a day. This includes only the process of separation and the process of reading the curve, sequencing reaction, cleaning and dissolving in a suitable buffer solution must be done separately. To obtain labelled reaction results that can be detected from printed DNA, the method of “cycle sequencing” is most commonly used. In this method are carried out successively primary annealing, extension by DNA polymerase and denaturation (melting) strands of printed DNA repeatedly (25-40 rounds). The main advantage of cycle sequencing is that it is more efficient. Each step in cycle sequencing is carried out by changing the reaction temperature using a thermal cycler PCR [5].
Topographically, the average sampling location is at an altitude of 800 meters above sea level. Judging from the climate, there is still a tropical climate which generally consists of five months of the rainy season and seven months of the dry season, and the air condition is quite fresh and clean because the pollution factor is still relatively low. The average annual rainfall is between 2000 and 2500 mm, while the average minimum air temperature is 24 0C and a maximum of 32 0C with climate, rainfall and air temperature factors indicating moderate conditions so that the flora and fauna life is very good.
Detection of CVPDr fragment distribution by PCR on several types of citrus plants showed that there were differences between susceptible and resistant citrus plants. Vulnerable citrus plants showed positive results containing CVPD, but also showed positive results containing CVPDr, as explained in table 1.
| Table 1: Results of Analysis of Several Citrus Plants on the distribution of CVPD and CVPDr fragments. | |||
| No. | Type of Citrus | CVPD | CVPDr |
| 1 | C. nobilis var. Kintamani | + | + |
| 2 | C. grandis L.Besakih | + | + |
| 3 | C. grandis L.Kintamani | + | + |
| 4 | C. grandis L. Bedugul | + | + |
| 5 | C. nobilis var. Bedugul | + | + |
| 6 | C. reticulate var. Slayer Mangguh | + | + |
| 7 | C. reticulate var. Slayer Kintamani | + | + |
| 8 | C. reticulate var. Gianyar tangerine | + | + |
| 9 | C. reticulate var. Keprok Petang | + | + |
| 10 | C. nobilis var. Gianyar | + | + |
| 11 | C. reticulate var. Tangerines Karangasem | + | + |
| 12 | C. reticulate var. Keprok Bangli | + | + |
| 13 | C. nobilis var. Mangguh | + | + |
| 14 | C. nobilis var. Denpasar | + | + |
| 15 | C. reticulate var. Keprok Mangguh | + | + |
| 16 | C. reticulate var. Tangerines Besakih | + | + |
| 17 | C. nobilis var. Tabanan | + | + |
| 18 | C. nobilis var Pecatu | + | + |
| 19 | C. nobilis var. Payangan | + | + |
| 20 | Triphasia trifoliate | - | + |
| 21 | C. reticulate var. Slayer Buleleng | + | + |
| 22 | C. amblycarpa | + | + |
| 23 | C. aurantifolia var. Seedles | - | + |
| 24 | C. nobilis var. Buleleng | + | + |
| 25 | C. aurantifolia | + | + |
| 26 | C. nobilis var Petang | + | - |
| 27 | Murraya paniculata L. | + | - |
| Note: + (containing); and - (not containing). | |||
PCR analysis results showed that from 27 samples of citrus plants, as many as 25 samples contained DNA fragments of CVPDr (841 bp). However, only 2 citrus species are resistant to CVPD disease, namely T. trifoliate and C. aurantifolia var. Seedless citrus. On the other hand there are citrus plants that are prone to CVPD. The study also found that two samples from 27 citrus plants did not contain DNA fragments of CVPDr, namely C. nobilis var. Petang and M. paniculata L. PCR analysis results are presented in figures 1,2.
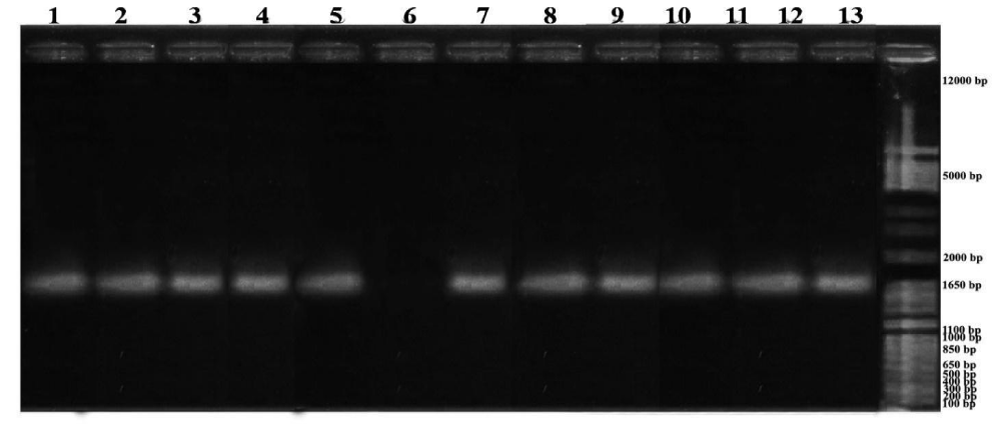
Figure 1: Citrus PCR Results: 1. C.grandis L.var. Kintamani, 2. C. nobilis var. Bedugul, 3. C. grandis L var. Bedugul, 4. C. reticulate var. Keprok Petang, 5. C. grandis L var. Besakih, 6. C. aurantifolia var. Seedles (not detected), 7. C. aurantifolia, 8. C. amblycarpa, 9. C. reticulate var Keprok Bangli, 10. C. reticulate var. Slayer Mangguh, 11. C. nobilis var. Mangguh, 12. C. reticulate var. Keprok Mangguh, 13. C. reticulate var. Keprok Besakih.
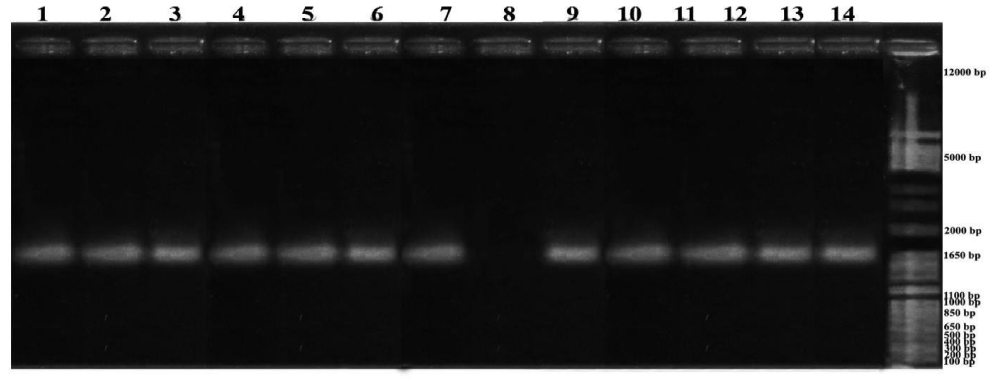
Figure 2: Results of PCR citrus plant samples: 1. C. nobilis var. Petang, 2. Muraya paniculate L. 3. C. reticulate var. Keprok Karangasem, 4. C. nobilis var. Kintamani, 5. C. nobilis var. Denpasar, 6. C. nobilis var. Buleleng, 7. C. nobilis var. Siam Gianyar, 8. T. trifoliata. (not detected), 9. C. nobilis var. Pecatu, 10. C. reticulate var. Buleleng Slayer, 11. C. nobilis var. Payangan, 12. C. reticulate var. Slayer Kintamani, 13. C. nobilis var. Tabanan, 14. C. reticulate var. Keprok Gianyar.
Research polymorphisms of genes CVPDr on some citrus plants using a sample of citrus plants in Bali, as a control used citrus T. trifoliata, and lemon without seeds (C. aurantifolia var. Seedless) which is known as a citrus plant which is resistant to CVPD attacks. To get the CVPDr gene polymorphism among several citrus plants in Bali sequencing was done on the PCR results using primers forward (1F): GACTAGGTGGTAATAACTACTTTT and primers reverse (1R): CCTTTTTGGTCTATCTTTACTTAG and base amplified from the PCR process were 841 bp. Total DNA isolation using the Mini Kit Plant from the nucleoSpin® Plant II from Marchery-Nagel. In the PCR reaction, was used 1 ng DNA samples, primers of 100 mole P011 and P012c, 2μl dNTP, 2μl PCR Buffer (10X), 0.2 Taq polymerase (5 U/μl). The PCR program uses Pre-treatment at 92ºC for 30 seconds with one rotation, denaturation at 92ºC for 60 seconds, annealing at 60ºC for 30 seconds, Elongation at 72ºC for 90 seconds with 40 turns. Extension at 72ºC for 90 seconds with one rotation [3]. PCR products were examined by electrophoresis in agarose gel TA 1% (w/v) and then squencing DNA fragments CVPDr. The sequencing process uses only primers forward (1F): GACTAGGTGGTAATAACTACTTTT and primers riverse (1R): CCTTTTTGGTCTATCTTTACTTA. PCR analysis results are presented in figures 3,4.
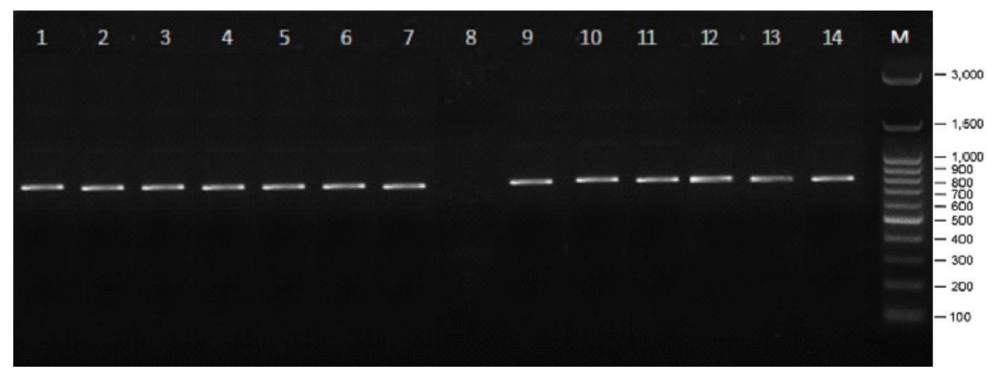
Figure 3: PCR results of citrus plant samples: 1. T. trifoliate, 2. C. aurantifolia var. Seedles, 3. C. reticulate var. Keprok Karangasem, 4. C. nobilis var Kintamani, 5. C. nobilis var Denpasar, 6. C. nobilis var Buleleng, 7. C. nobilis var Siam Gianyar, 8.C. nobilis var Petang (not detected), 9. C. nobilis var Pecatu, 10. C. reticulate var Buleleng Slayer, 11. C. nobilis var Payangan, 12. C. reticulate var Slayer Kintamani, 13. C. nobilis var Tabanan, 14. C. reticulate var Keprok Gianyar..
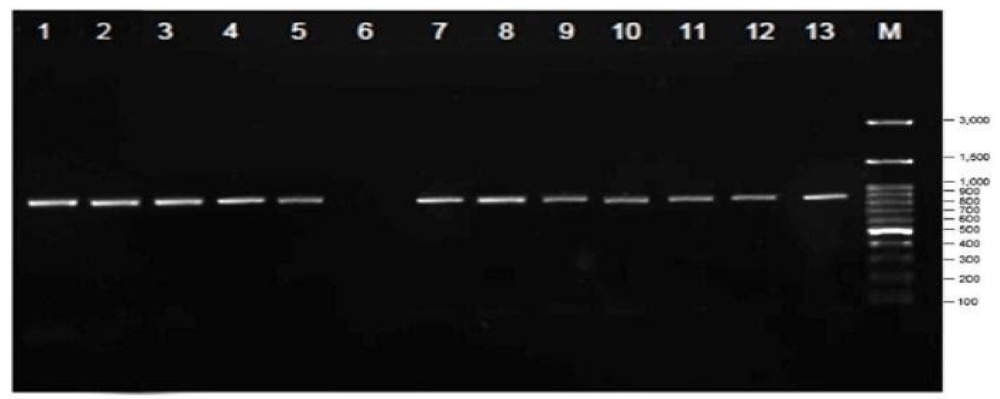
Figure 4: Citrus PCR Results: 1. C. grandis L var. Kintamani, 2. C. nobilis var. Bedugul, 3. C. grandis L var. Bedugul, 4. C. reticulate var. Keprok Petang, 5. C. grandis L var. Besakih, 6 . M. paniculata L. (not detected), 7. C. aurantifolia, 8. C. amblycarpa, 9. C. reticulate var. Keprok Bangli, 10. C. reticulate var. Slayer Mangguh, 11. C. nobilis var. Mangguh, 12. C. reticulate var. Keprok Mangguh, 13. C. reticulate var. Keprok Besakih.
PCR analysis of DNA isolated from samples of several resistant and susceptible citrus plants showed that from 27 citrus plants samples of 25 samples contained CVPDr (resistance genes), this was evidenced by their presence at 841 bp. The primary sequence used is the primer for CVPDr detection. In figure 3a sample of the Siam Petang (C. nobilis var. Petang) not detected in line 4 (four), this proves that the sample of the plant did not have a resistance gene, it is likely that the sample used had a severe CVPD disease and was not detected by using the primary resistance gene.
In another PCR analysis that is in figure 4 shows that Kemuning (M. peniculata) on line 6 was not detected so there was no CVPDr gene. This is because M. peniculata is a host plant for fleas. CVPD infectious disease insects will be more active at high temperatures (lowlands) compared to low temperatures (highlands). From the results of this study found the presence of CVPDr gene in all types of citrus both vulnerable and resistant such as Tangerines, Siem citrus, Selayar citrus, Balinese citrus, Kingkit citrus and Seedless Lime citrus in all samples.
Polymorphism of DNA Fragments of CVPDr genes
In addition, 25 of 27 citrus plants sample from the study showed that the sample contained DNA fragments of CVPDr, susceptible to CVPD disease. The results of this study indicate that the CVPDr DNA fragment in this citrus species or varieties does not work well and can be caused by mutations or resistance to CVPD disease needed by other genes. Therefore, we study the CVPDr DNA fragment polymorphism. The DNA sequence DNA fragment CVPDr is then performed and the results are shown in table 2.
| Table 2: CVPDr gene polymorphism in some citrus plants in Bali. | |||
| No. | Size of bases (bp) | Bases Polymorphism | Isolate Sample |
| 1 | 117 | TCA TCT | CnKin |
| 2 | 145-147 | GAG TAA | Tt, CavarSeed, CnBll, CnPc, CnvarSlayBll, CnPy, CnTbn, CrvarKepMang, CrvarKepBs. |
| 3 | 149 | CTT CGT | CnKin |
| 4 | 166-167 | TGG CAG | CgKin, CgBs |
| 5 | 181 | AAA GAA | CgKin, CgBd |
| 6 | 196 | CAA TAA | CnKin |
| 7 | 247 | GGG AGG | CnKin |
| 8 | 264 | CAGCAT | CrvarSlayKin, CrvarKepGr , CrvarSlayMang, CrvarKepPet |
| 9 | 269 | TAT TGT | CnKin, CnBd, CgKin, CgBd, CgBs |
| 10 | 275 | AAT ACT | CnKin, CnBd, CgBs, |
| 11 | 278 | GAC G · C | CnKin |
| 12 | 298 | AGC GGC | CnBd |
| 13 | 315 | TAG TAT | CnBd, CnKin, CgBd, CgBs |
| 14 | 336 | AAA AAG | CnKin |
| 15 | 344 | GTT GCT | CnKin |
| 16 | 347 | TAT TCT | CrvarSlayKin, CrvarKep, CrvarSlayMang, CrvarKepPet |
| 17 | 352 | AAT CAT | CnKin |
| 18 | 400-424 | AGACTTTGTAGGCATAAATGCTAT | Only CnKin mutated |
| 19 | 462 | TTT TTG | CgBd, CrvarKepBgl |
| 20 | 478 | AAA CAA | CgBs |
| 21 | 490 | CAG TAG | CgKin |
| 22 | 503-504 | TTG AAG | CnKin, CnBd, CgKin, CgBd, CnBll, |
| 23 | 527 | GTT CTT | CnBd, |
| 24 | 538 | ATT ATG | CnKin, CnBd, CgKin, CgBd, CnBll |
| 25 | 575 | TAA CAA | CnKin |
| 26 | 580-841 | NNN NN | CrvarKepGr |
| 27 | 585 | TCT TTT | CnKin, CnKin, CnKin, CnKin, CnKinNNN NN CNPC, CnTbn, CnBd |
| 28 | 591 | TGG TAG | CnKin, CnBd, CgBd, CgKin, CgBs |
| 29 | 597 | ACC ATC | CnKin, CnBd, CgBd, CgKin, CgBs |
| 30 | 631 | TTC TTA | CnKin |
| 31 | 633 | ATA AGA | CnKin , CrvarSlayKin, CnBd, CgKin, CgBd, CrvarKepMang, CrvarKepBs, CgBs, CrvarKepBgl |
| 32 | 652 | ATG ATC | CrvarKepBgl, CgBs |
| 33 | 663 | AAA ACA | CnKin, CnBd, CgKin, CgBd, CgBs |
| 34 | 672 | TGT TTT | CgBs |
| 35 | 680 bp | A · T ATT | CrvarSlayMang |
| 680 bp | A · T AAT | CnBll | |
| 680-841 | Bases there is a difference with another sample | CrvarSlayMang | |
| 36 | 694 | TTC TTG | CnBd, CgKin, CgBd |
| 37 | 706 | AA · AAC | CnTbn |
| 38 | 709 | GTC GTT | CgBs, CnKin |
| 39 | 714 | TTA TCA | CnKin |
| 40 | 724 | ATA ATC | CgKin, CgBd |
| 41 | 747 | ATA AGA | CnBd |
| 42 | 783 | ATA AGA | CnBd |
| 43 | 801 | T · A TTT | CnKin |
| Note: CnKin = C. nobilis var. Kintamani, Tt =T. trifoliata, CavarSeed = C. aurantifolia var. Seedles, CnBll = C. nobilis var. Buleleng, CnPc = C. nobilis var. Pecatu, Crvar.SlayBll = C. reticulate var. Slayer Buleleng, CnPay = C. nobilis var. Payangan, CnTab = C. nobilis var. Tabanan, CrvarKepMang = C. reticulate var Keprok Mangguh, CrKepBs = C. reticulate var. Keprok Besakih, CrvarSlayMng = C. reticulate var. Slayer Mangguh, Cauran = C. aurantifolia, Cambly = C. amblycarpa, CgKin = C. grandis C. L. Kintamani, CgBs = C. grandis L. var. Besakih, CgBd =C. grandis L.var. Bedugul, CrvarSlayKin =C. reticulate var. Slayer Kintamani, CrvarKepGr =C. reticulate var. Keprok Gianyar, CrvarKepPet =C. reticulate var. Keprok Petang, CnBd =C. nobilis var. Bedugul, CnGr =C. nobilis var. Gianyar, CrvarKepKr =C. reticulate var. Keprok Karangasem, CrvarBgl =C. reticulate var. Keprok Bangli, CnMang =C. nobilis var. Mangguh, CnDps =C. nobilis var. Denpasar | |||
Results showed a CVPDr gene polymorphism between Tangerines citrus, Siam citrus, Selayar citrus, T. trifolia, Seedles Lime citrus, and all the citrus that are prone to CVPD. The occurrence of polymorphism in a gene, indicates the existence of some differences in DNA sequence in citrus plants sampled with different or similar species. The difference in DNA sequence is caused by the occurrence of deletions, insertions, recombination, low random marriage and selection in the population [6]. So far, through library research there has not been found a research report on the polymorphism of the CVPDr gene fragment in citrus plants in Indonesia. In this study the CVPDr gene showed that the target band was detected positively in 25 samples from 27 samples studied at 841 bp. The electrophoresis method is a general method that can be applied to show polymorphisms of a large number of enzymes. With this method, it can be found changes in the structure of enzyme molecules that cause changes in the molecular electrical charge [7].
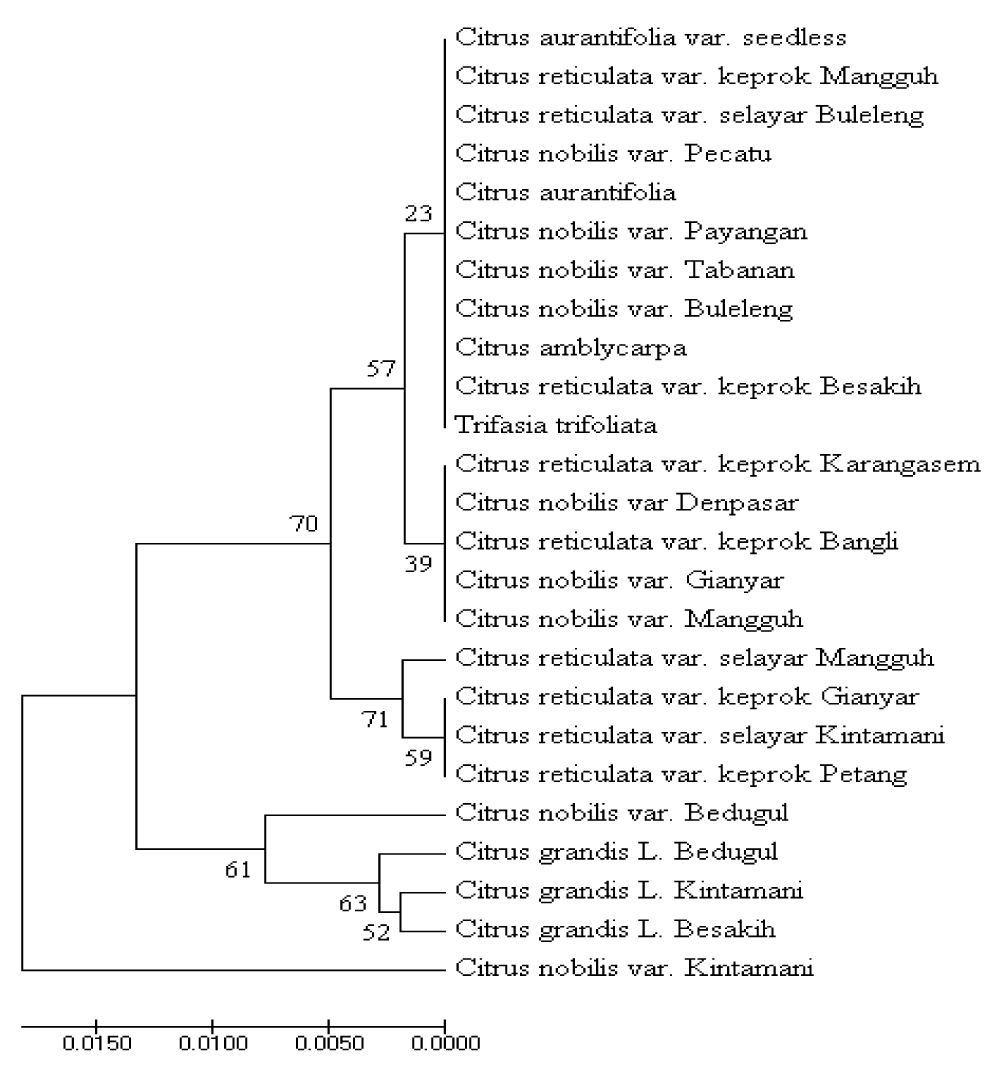
Figure 5 shows the phylogeny of several species of citrus plants in Bali. The phylogenetic tree is a logical approach to show the evolutionary relationship between organisms. Phylogenetics is a model to represent the relationship between ancestral organisms, molecular sequences or both. One of the goals of phylogenetic compilation is to precisely construct relationships between organisms and estimate differences that occur from an ancestor to its offspring. Phylogenetics can analyzed the changes that occur in the evolution of different organisms. Based on the analysis, sequences that have closeness can be identified by occupying branches next to the tree.
The results of this study indicate that there are many DNA polymorphisms among samples in the CVPDr DNA fragment. Phylogenetic trees derived from sample polymorphisms show that Kingkit citrus (T. trifoliate var. Seedless) and Lime citrus (C. aurantifolia) are located in the same cluster, while other samples are in different clusters. These results indicate that DNA polymorphisms in CVPDr DNA fragments cause differences in the resistance of citrus plants to CVPD disease. Polymorphism is the presence of more than one allele at a genetic locus with an allele frequency rarely more than one percent in the population. Gene polymorphisms occur due to changes in the composition of the nucleotides of genes. Changes in gene composition are influenced by several factors such as natural or artificial selection, mating and mutation. These changes can affect an organism’s phenotypic changes [8].
Kinship and evolutionary relationships between organisms can be described through phylogenetic trees. The purpose of studying phylogenetic trees is to reconstruct genealogical ties (genealogies) between organisms and determine the evolutionary time of an organism. Phylogenetic tree is a graph consisting of branches or nodes. Two adjacent nodes are connected by one branch. Nodes represent taxonomic units, while branches represent relationships between taxonomic units. Branching patterns on phylogenetic trees are called topologies. The length of a branch represents the difference in that branch. Taxonomic units can be species, populations, individuals or genes [9].
Phylogenetic trees can be reconstructed using several statistical methods. One of them is the statistical method that is commonly used, namely Bayesian methods. Bayesian method is a phylogenetic tree reconstruction method that uses optimality criteria (optimal standard). The method has the same concept as the likelihood method, which uses probability distribution of trees to find the most appropriate phylogenetic tree from the data sequences used. The position of each sequence in aligned sequences is called character, while nucleotides are called “states” in the Bayesian methods. The position of all characters is analyzed independently or separately from each other, so that each alignment column is assumed to be an independent realization in the evolutionary process. Bayesian methods allow researchers to determine their own models such as the model parameter substitution, long branches, and tree topology [10], as shown in figure 5.
Figure 5: Phylogenetic tree is the result of UPGMA analysis by Bootstrapping various types of citrus plants in several regions in Bali based on nucleotide sequences. The length of each pair of the branches above shows the distance between each pair.
There are fragment DNA polymorphism CVPDr on citrus that vulnerable and citrus plants that are resistant or tolerant to CVPD disease. PCR test showed T. trifoliata and C. aurantifolia var. Seedless are resistant to CVPD, and Pylogenically are in the same group as C. nobilis var. Buleleng, C. nobilis var Tabanan, C. nobilis var Payangan, C. nobilis var Pecatu, C. reticulate var. Slayer Buleleng, C. reticulate var. Keprok Manggu, C. reticulate var. Keprok Besakih, and C. amblycarpa. On the other hand, citrus plants that are sensitive to CVPD are in a different group. There are two types of citrus plants not containing CVPDr DNA fragments, namely C. nobilis var. Petang and M. paniculata L. CVPDr DNA fragments indicated have an important role in the mechanism of resistance of citrus plants to CVPD disease.
This study was supported by Udayana University Research Grant No.: 383-5/UN14.4.A/LT/2018
- Wahyuningsih E. CVPD on Citrus (Citrus spp) and Control Efforts. Jakarta: Vis Vitalis. 2009; 2:
- Mahayani. AAPS. Analysis of Gene Expression Clones CVPDr In Escherichia coli cells. Surabaya: Agroknow J. 1:
- Wirawan I GP, Liliek S, Wijaya. CVPD Disease in Citrus Plants. Denpasar: Udayana University Press.2004.
- Wirawan I GP, Julyasih SM, Adiartayasa W, Wijaya N, Anom P. Increasing Local Fruits Competitiveness in Entering The Tourism Market in Bali. Int J Biosciences Biotechnol. 2014; 2:
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. USA: Proc Natl Acad Sci USA. PubMed: 1977; 74: 5463-5467. https://www.ncbi.nlm.nih.gov/pubmed/271968
- Schleif R. Genetics and Molecular Biology 2 years. The John Hopkins Press Ltd. London. 1993.
- Harris H. Fundamentals of Human Biochemical Genetics. Third Edition. Published ASM Sofro. Gadjah Mada University Press. Yogyakarta. 1994.
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. Cambridge University Press. 2002.
- Li WH. Molecular evolution, Sinauer Associates, Inc., Publisher, Sunderland Massachusetts. 1997.
- Vandamme AM. Basic concepts of molecular evolution. In: Lemey P, Salemi M, Vandamme AM. (ed). The phylogenetic. 2009.